PBANKA_0517500 bromodomain protein 1, putative (BDP1)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. falciparum 3D7 | Asexual |
Refractory |
26067602 (Conditional) | Theo Sanderson, Wellcome Trust Sanger Institute |
| P. falciparum 3D7 | Asexual |
Difference from wild-type |
26067602 (Conditional) | Theo Sanderson, Wellcome Trust Sanger Institute |
Imaging data (from Malaria Metabolic Pathways)
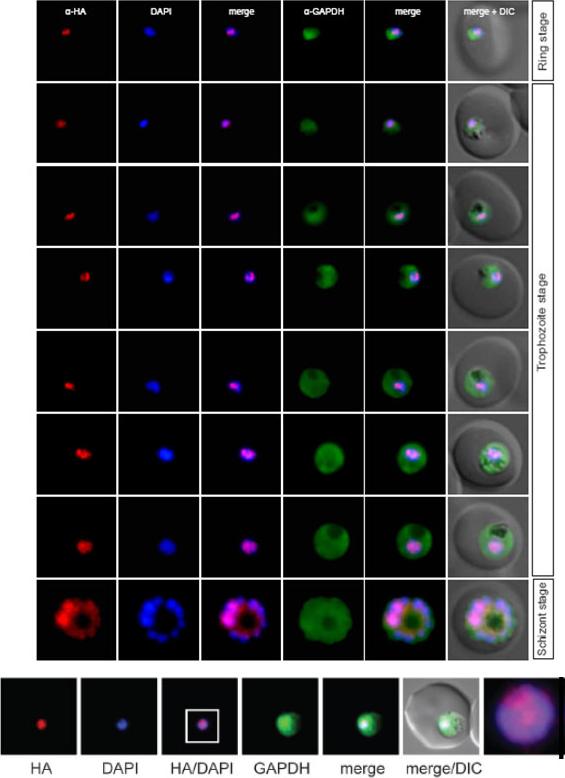
Localisation of PF10_0328 during the IDC. Localisation of the tagged protein was visualised using anti-HA antibodies (red). Antibodies against GAPDH were used to visualise the cytosolic compartment. DAPI was used to visualise the nucleus. DIC images are shown as reference. The protein localized to unknown nuclear subcompartment.Oehring SC, Woodcroft BJ, Moes S, Wetzel J, Dietz O, Pulfer A, Dekiwadia C, Maeser P, Flueck C, Witmer K, Brancucci NM, Niederwieser I, Jenoe P, Ralph SA, Voss TS. Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol. 2012 Nov 26;13(11):R108.
See original on MMP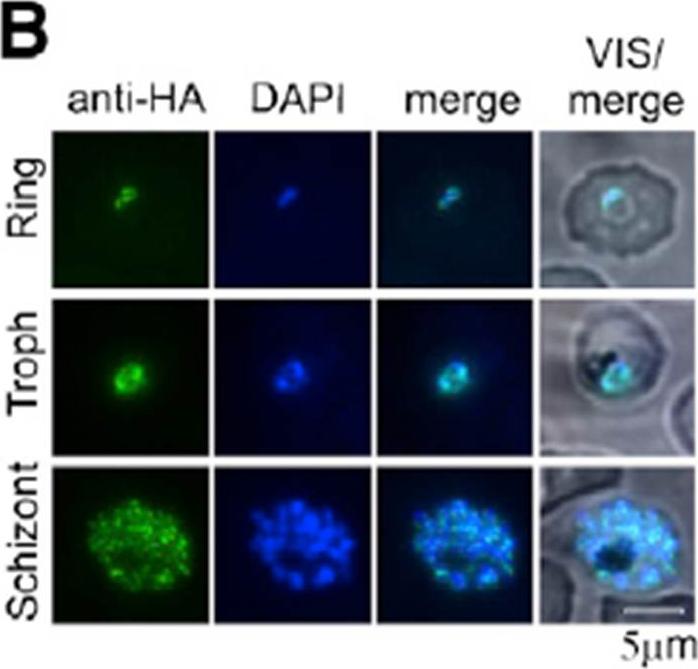
Immunofluorescence detection of PfBDP1HA in the nucleus across the asexual life cycle of P. falciparum.Josling GA, Petter M, Oehring SC, Gupta AP, Dietz O, Wilson DW, Schubert T, Längst G, Gilson PR, Crabb BS, Moes S, Jenoe P, Lim SW, Brown GV, Bozdech Z, Voss TS, Duffy MF. A Plasmodium Falciparum Bromodomain Protein Regulates Invasion Gene Expression. Cell Host Microbe. 2015 17(6):741-51.
See original on MMPMore information
| PlasmoDB | PBANKA_0517500 |
| GeneDB | PBANKA_0517500 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PB001232.02.0, PB001419.02.0, PB301031.00.0, PBANKA_051750 |
| Orthologs | PCHAS_0517600 , PF3D7_1033700 , PKNH_0618500 , PVP01_0618800 , PVX_111040 , PY17X_0518600 |
| Google Scholar | Search for all mentions of this gene |