PVX_111065 early transcribed membrane protein (ETRAMP)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
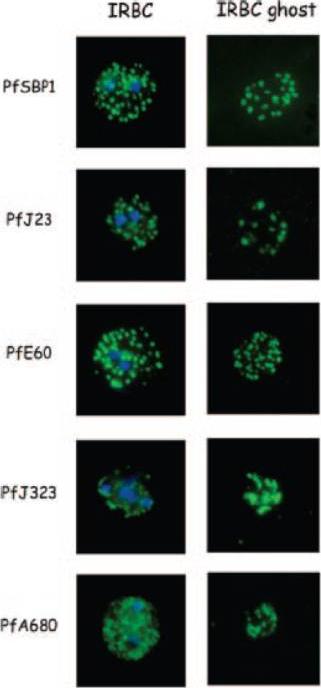
Indirect immunofluorescence of P. falciparum-infected erythrocytes and infected red blood cell (IRBC) ghosts. Air-dried infected red blood cells and infected red blood cell ghosts were incubated with mouse antibodies raised against GST fusion proteins as indicated. The nuclei were stained with DAPI (blue). Negative controls were performed using preimmune sera and anti-GST antibodies (not shown). All proteins are associated with Maurer;s clefts.Vincensini L, Richert S, Blisnick T, Van Dorsselaer A, Leize-Wagner E, Rabilloud T, Braun Breton C. Proteomic analysis identifies novel proteins of the Maurer's clefts, a secretory compartment delivering Plasmodium falciparum proteins to the surface of its host cell. Mol Cell Proteomics. 2005 4:582-93.
See original on MMP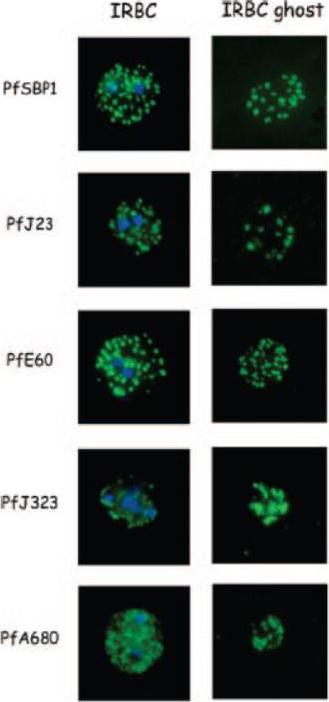
Indirect immunofluorescence of P. falciparum-infected erythrocytes and infected red blood cell (IRBC) ghosts. Air-dried infected red blood cells and infected red blood cell ghosts were incubated with mouse antibodies raised against GST fusion proteins as indicated. The nuclei were stained with DAPI (blue). Negative controls were performed using preimmune sera and anti-GST antibodies (not shown). All proteins are associated with Maurer;s clefts.Vincensini L, Richert S, Blisnick T, Van Dorsselaer A, Leize-Wagner E, Rabilloud T, Braun Breton C. Proteomic analysis identifies novel proteins of the Maurer's clefts, a secretory compartment delivering Plasmodium falciparum proteins to the surface of its host cell. Mol Cell Proteomics. 2005 4:582-93.
See original on MMPMore information
| PlasmoDB | PVX_111065 |
| GeneDB | PVX_111065 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | Pv111065 |
| Orthologs | PF3D7_1033200 , PKNH_0618000 , PVP01_0618300 |
| Google Scholar | Search for all mentions of this gene |