PVX_092040 geranylgeranyl pyrophosphate synthase (GGPPS)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
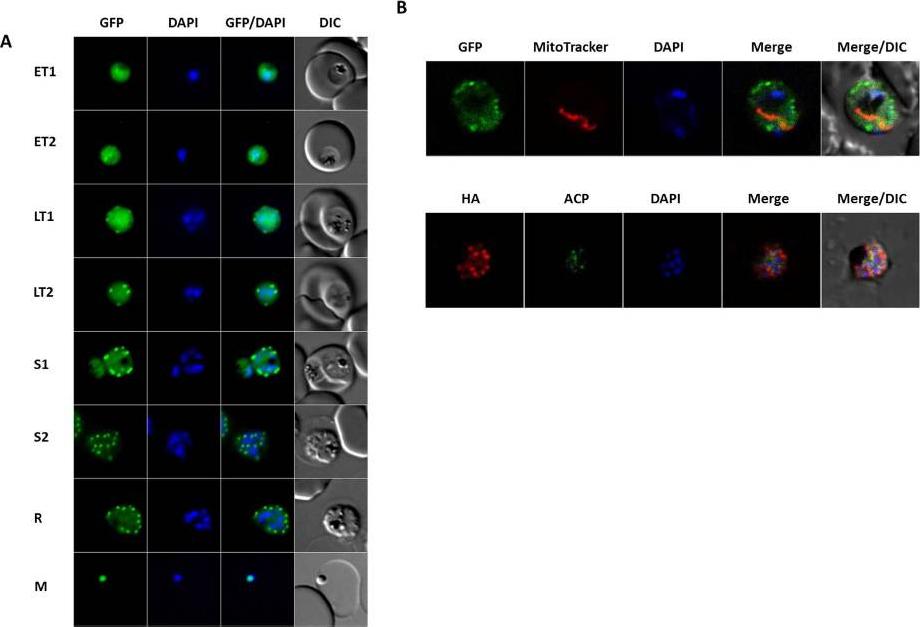
FPPS/GGPPS localization. Images of live parasites (FPPS/GGPPS-HA-GFP strain) expressing GFP and immunofluorescence of the FPPS/GGPPS-HA strain visualized by fluorescence or confocal microscopy. (A) Images of live parasites FPPS/GGPPS-HA-GFP by fluorescence microscopy during the intra-erythrocytic cycle of the parasite. (B) Bottom -immunofluorescence of the FPPS/GGPPS-HA strain with antibody as indicated, visualized by confocal microscopy. Top—Images of live FPPS/GGPPS-HA-GFP parasites with markers as indicated, visualized by confocal microscopy. ET1 (early rophozoites); ET2 (early trophozoites); LT (latetrophozoites); LT (late trophozoites) S1 (schizont); S2 (schizont); R (rosettes); M (Merozoites); DAPI (nucleus marker). Original magnification for all images: 1.000×Gabriel HB, de Azevedo MF, Palmisano G, Wunderlich G, Kimura EA, Katzin AM, Alves JM. Single-target high-throughput transcription analyses reveal high levels of alternative splicing present in the FPPS/GGPPS from Plasmodium falciparum. Sci Rep. 2015. PMID:
See original on MMPMore information
| PlasmoDB | PVX_092040 |
| GeneDB | PVX_092040 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | Pv092040 |
| Orthologs | PBANKA_0919800 , PCHAS_0924600 , PF3D7_1128400 , PKNH_0926500 , PVP01_0929200 , PY17X_0921900 |
| Google Scholar | Search for all mentions of this gene |