PVP01_1221000 histone H3-like centromeric protein CSE4, putative (CenH3)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
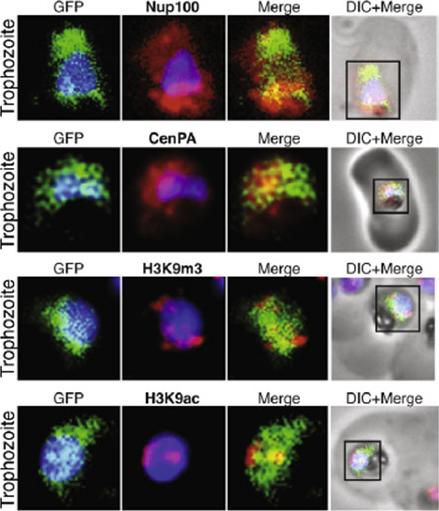
The protein encoded by the gene MAL13.P1.122 localize to the far nuclear periphery. MAL13P1.122 is a SET-domain protein (Histone-lysine N-methyltransferase), which localizes to the nuclear periphery surrounding the DAPI stained portion within the same region of CenPA (histone H3) and the histone mark H3K9m3. Co-localization studies by immunofluorescence. Nup100 (PFI0250c) and CenPA (histone H3 PF13_0185) are used as markers of the nuclear periphery.Volz J, Carvalho TG, Ralph SA, Gilson P, Thompson J, Tonkin CJ, Langer C, Crabb BS, Cowman AF. Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum. Int J Parasitol. 2010 40:109-21. Copyright Elsevier 2010.
See original on MMP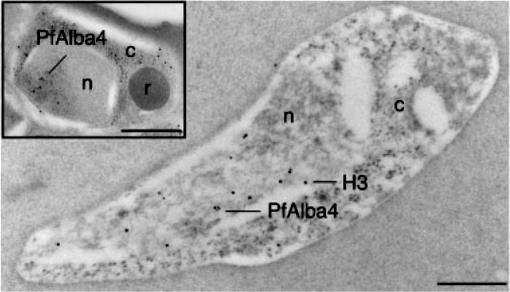
Immuno-EM was performed on ring stage parasites using anti-PfAlba4 antibodies (Ab 237–2) bound to 10nm gold particles and anti-H3 antibodies bound to 15nM gold particles. The inset shows a single developing merozoite within schizont-stage parasites labeled with anti-PfAlba4 antibodies (Ab 237–2) bound to 10nm gold particles; n=nucleus, c=cytoplasm, r=rhoptry; Scale bars represent 500 nm. In the ring stage, we detected PfAlba4 uniquely in the nucleus. Since the nuclear membrane is difficult to visualize in immuno-EM in the ring stage, co-labeling with antibodies directed against histone H3 was used to determine the area of the nucleus in this parasite form. The images localize PfAlba4 at the nuclear periphery. This pattern was confirmed in developing merozoites (inset) where distinct clusters of PfAlba4 could be detected at the periphery of the nuclei.Chêne A, Vembar SS, Rivière L, Lopez-Rubio JJ, Claes A, Siegel TN, Sakamoto H, Scheidig-Benatar C, Hernandez-Rivas R, Scherf A. PfAlbas constitute a new eukaryotic DNA/RNA-binding protein family in malaria parasites. Nucleic Acids Res. 2011 Dec 13. [Epub ahead of print]
See original on MMP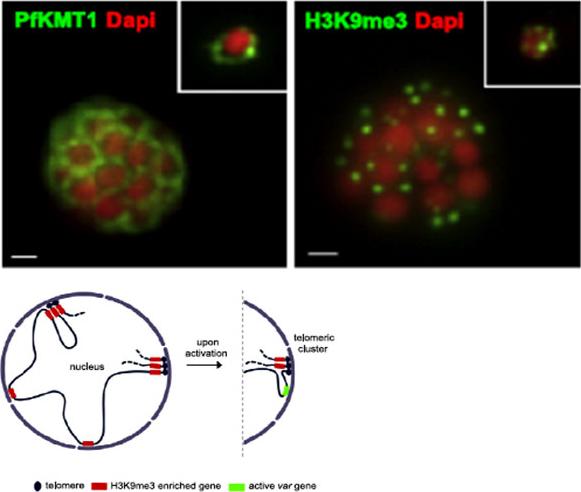
PfKMT1 and genes enriched in H3K9me3 are positioned at the nuclear periphery. Immuno-fluorescence assays using antibodies against H3K9me3 H3 (right) and the P. falciparum putative H3K9 methyltransferase PfKMT1 (left). 3D7 parasites are in late schizont (main image) and ring stage (top-right square). Nuclear DNA was detected with DAPI (red).Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009 5:179-90.
See original on MMP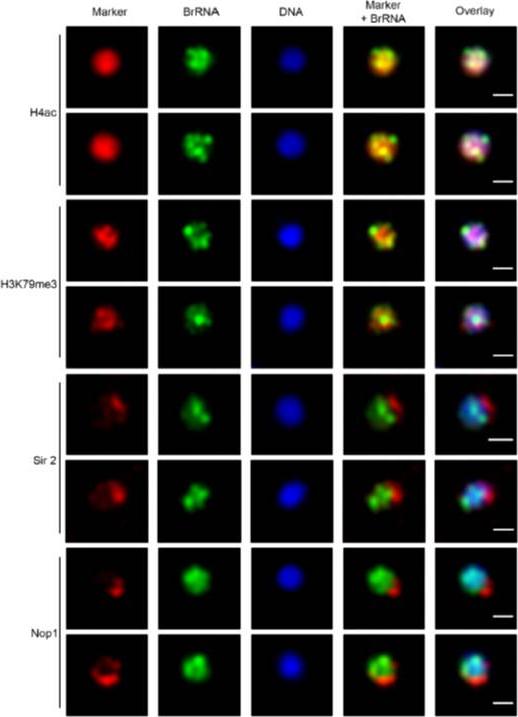
Transcription sites form a distinct compartment in the nucleus of P. falciparum. Immunofluorescence assay for nascent RNAs of ring stage parasites labeled with BrUTP (BrRNA, green) and the compartments of acetylated H4 (H4ac, active chromatin); a proposed histone marker of nascent transcription (H3K79me3; PfSir2A (silencing compartment); and Nop1 (nucleolar compartment). DNA was labeled by DAPI, in blue. Bars, 1 mm.Moraes CB, Dorval T, Contreras-Dominguez M, Dossin Fde M, Hansen MA, Genovesio A, Freitas-Junior LH. Transcription sites are developmentally regulated during the asexual cycle of Plasmodium falciparum. PLoS One. 2013;8(2):e55539.
See original on MMP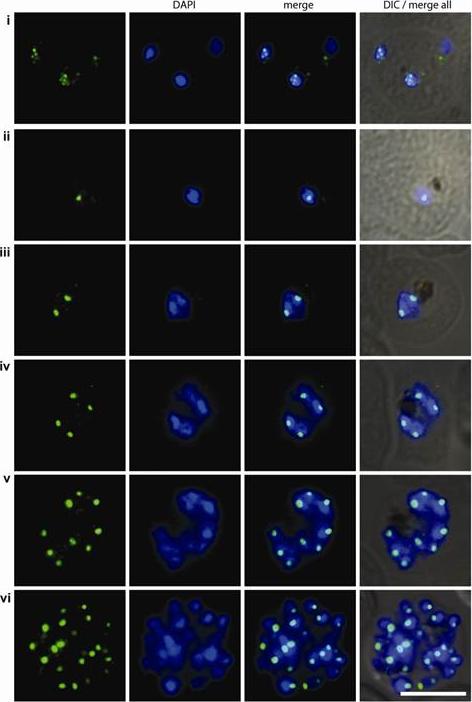
Hoeijmakers WA, Flueck C, Françoijs KJ, Smits AH, Wetzel J, Volz JC, Cowman AF, Voss T, Stunnenberg HG, Bártfai R. Plasmodium falciparum centromeres display a unique epigenetic makeup and cluster prior to and during schizogony. Cell Microbiol. 2012 14(9):1391-401.
See original on MMP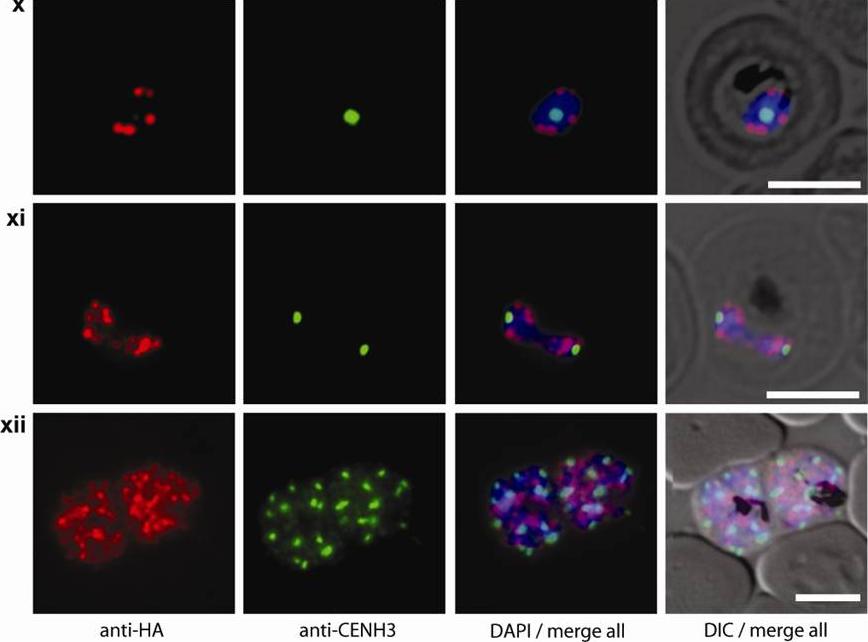
Double staining immunofluorescence pictures obtained from the transgenic parasite line 3D7/HP1-HA expressing HA-tagged PfHP1. Cells were simultaneously probed with aHA and aPfCENH3 antibodies. PfCENH3 foci do not overlap with PfHP1-marked chromosome end clusters. GFP live fluorescence and aPfCENH3 6 antibody (green), aHA antibody (red), DAPI nuclear stain (blue), DIC (differential interference contrast). Scale bars are 5 μm long.Hoeijmakers WA, Flueck C, Françoijs KJ, Smits AH, Wetzel J, Volz JC, Cowman AF, Voss T, Stunnenberg HG, Bártfai R. Plasmodium falciparum centromeres display a unique epigenetic makeup and cluster prior to and during schizogony. Cell Microbiol. 2012 14(9):1391-401.
See original on MMP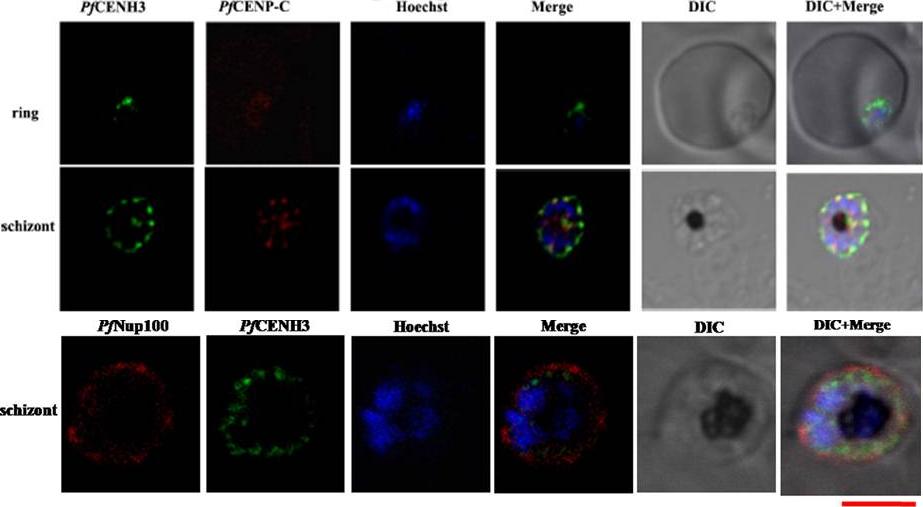
Upper: Colocalization of PfCENH3 with PfCENP-C. Confocal images showing the co-localization of the putative PfCENP-C (red) and PfCENH3 (green) at the nuclear periphery. The nucleus is stained with Hoechst (blue). Scale bar = 5 mm. Lower: The fluorescence pattern obtained on merging PfCENH3 and PfNup100with the nucleus depicts that PfCENH3 localizes to the nuclear periphery just outside the Hoechst stained nucleus but well within the PfNup100 that marks the outer nuclear boundaries.Verma G, Surolia N. Plasmodium falciparum CENH3 is able to functionally complement Cse4p and its, C-terminus is essential for centromere function. Mol Biochem Parasitol. 2013 Dec 6. [Epub ahead of print]
See original on MMP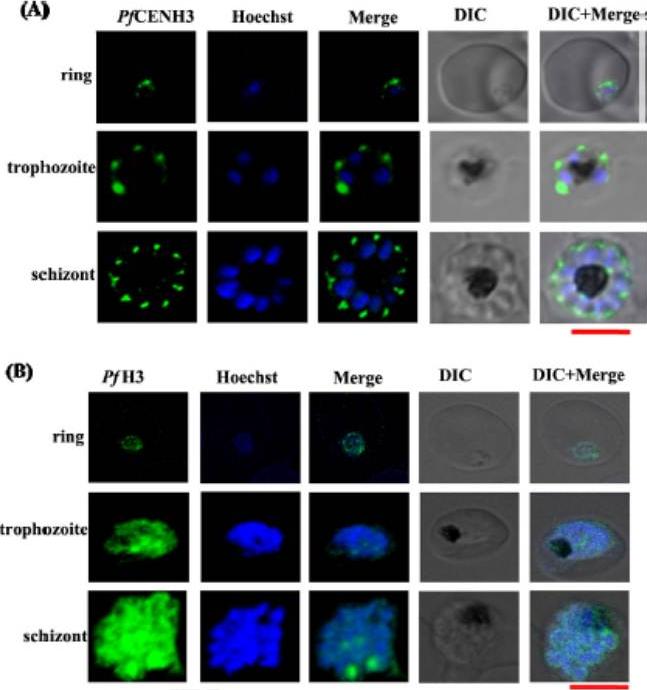
PfCENH3 is expressed at the perinuclear regions while PfH3 localizes to nucleus. (A, B) The confocal images for immuno-fluorescence assays represent the subcellular localization pattern of PfCENH3 and PfH3 (green) with respect to the Hoechst stained nucleus (blue) at ring, trophozoite and schizont stages of P. falciparum respectively. While PfH3 localizes throughout the nuclear mass, PfCENH3 localizes towards perinuclear regions at various intraerythrocytic stages. At ring stage, PfCENH3 localizes as a single cluster per nucleus. At trophozoite and schizont stages it localizes as discrete spots on each nucleus. Scale bar = 5 mm.Verma G, Surolia N. Plasmodium falciparum CENH3 is able to functionally complement Cse4p and its, C-terminus is essential for centromere function. Mol Biochem Parasitol. 2013 Dec 6. [Epub ahead of print]
See original on MMP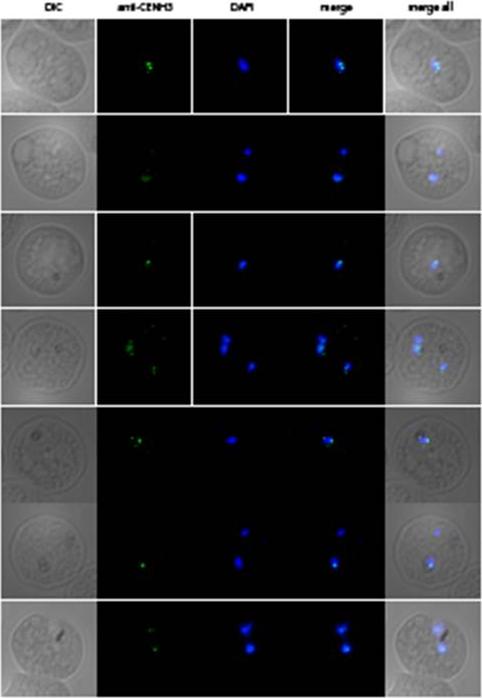
PfCENH3 localization in ring stage parasites. 3D7 wild-type parasites of ring stages were subjected to immunofluorescence assay with αPfCENH3. At this stage the PfCENH3 signal is substantially weaker and seldom concentrated to a single focus, suggesting that centromeres do not congregate into a single cluster at this stage. 3-6 distinct foci could typically be observed per nuclei potentially indicating sub-clustering of centromeres in ring stage nuclei.Hoeijmakers WA, Flueck C, Françoijs KJ, Smits AH, Wetzel J, Volz JC, Cowman AF, Voss T, Stunnenberg HG, Bártfai R. Plasmodium falciparum centromeres display a unique epigenetic makeup and cluster prior to and during schizogony. Cell Microbiol. 2012 14(9):1391-401.
See original on MMP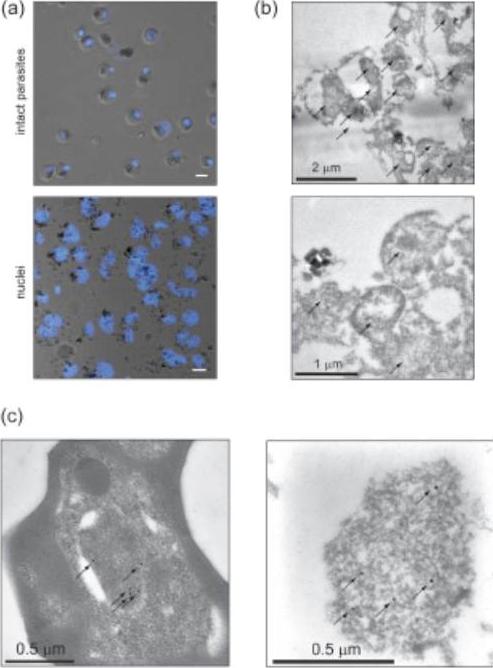
Preparation of crude P. falciparum nuclei for proteomic analysis. (a) Visual assessment of the crude nuclear preparation. Intact parasites (control) and isolated nuclei were stained with DAPI and analysed by DIC and fluorescence microscopy. Scale bar 2 mm. (b) TEM analysis of isolated nuclei shows intact as well as damaged nuclei and very little contamination from other organelles. Arrows indicate individual nuclei. (c) Immunoelectron microscopy verifies the identity of nuclei. Anti-H3K4me3 antibodies were used to label nuclei in intact cells (left panel) and in the nuclear preparation (right panel) (arrows). Oehring SC, Woodcroft BJ, Moes S, Wetzel J, Dietz O, Pulfer A, Dekiwadia C, Maeser P, Flueck C, Witmer K, Brancucci NM, Niederwieser I, Jenoe P, Ralph SA, Voss TS. Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol. 2012 Nov 26;13(11):R108.
See original on MMP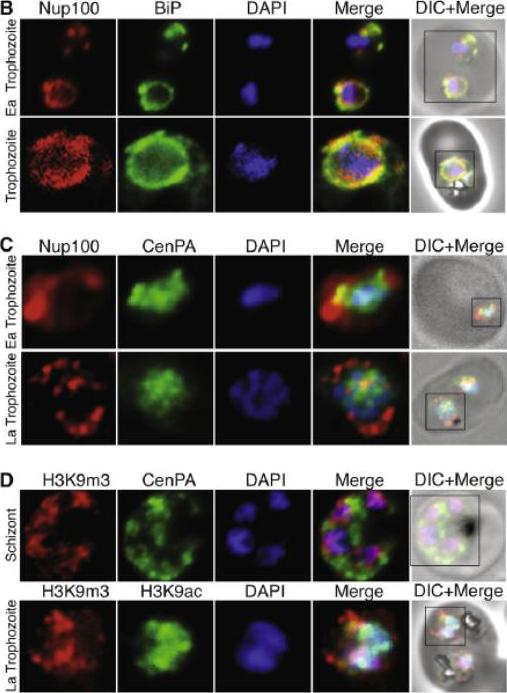
(B) Nucleoporin 100 (Nup100), a protein of the nuclear pore and BiP immunofluorescence analysis reveals the localization of Nup100 outside the DAPI stained area. Nup100 localizes in close proximity to BiP, a marker for the endoplasmic reticulum (ER) and does not overlap with DAPI, confirming that it defines the outer limits of the nucleus. (C) Localization of Nup100 and CenPA (histone 3) outside the DAPI stained area. CenPA and Nup100 do not co-localize; however, CenPA extends beyond the DAPI staining area of the nucleus suggesting that it is located within the periphery but inside the Nup100 delimited area of the nucleus (D) Localization of the histone marks H3K9m3 with CenPA and H3K9ac in the nucleus. The histone modification H3K9m3 silences transcription whereas H3K9ac is found associated with active transcription in general. H3K9m3 localized mostly to the nuclear periphery outside the DAPI stained area, which was consistent with the location of chromosome clusters containing silenced var genes. H3K9ac localized more with the DAPI stained area of the nucleus suggesting the DAPI stained area is broadly the transcriptionally active zone.Ea Trophozoite, Early Trophozoite; La trophozoite, Late Trophozoite.Volz J, Carvalho TG, Ralph SA, Gilson P, Thompson J, Tonkin CJ, Langer C, Crabb BS, Cowman AF. Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum. Int J Parasitol. 2010 40:109-21. Copyright Elsevier 2010.
See original on MMP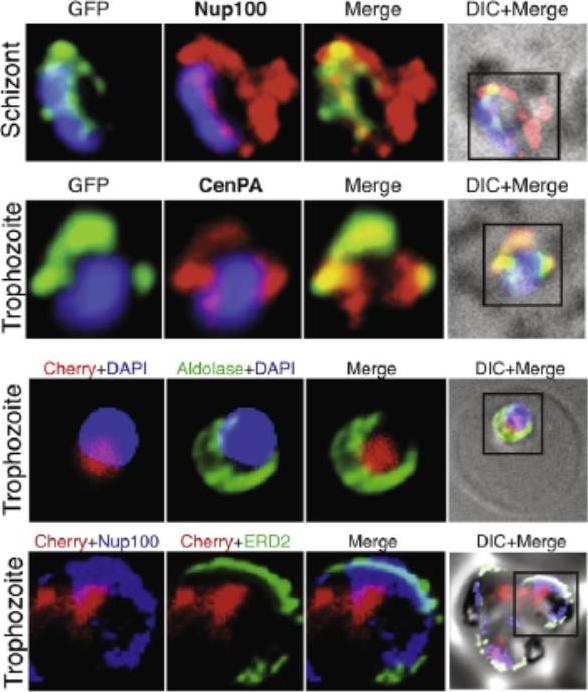
Nucleosome assembly protein PFI0930c localizes to the far nuclear periphery. (A,B) PFI0930c is either tagged with GFP or Cherry, respectively. Co-localization studies by immunofluorescence reveal its localization to the nuclear periphery, which is marked by CenPA (histone H3)and delimited by the nuclear envelope marker Nup100 (A), but not to the cytoplasmic portion of the cell marked by aldolase (PF14_0425) and ERD2 (PF13_0280) (B). Co-localisation of PFI0930c-cherry and aldolase, used as a cytoplasmic marker showed clear separation of nuclear and cytoplasmic regions in the P. falciparum cell. Co-localisation analysis with Nup100 and ERD2, a marker for the cis- and trans-Golgi showed the same overlapping localisation of PFI0930c-cherry and Nup100, both in an internal location relative to ERD2.Volz J, Carvalho TG, Ralph SA, Gilson P, Thompson J, Tonkin CJ, Langer C, Crabb BS, Cowman AF. Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum. Int J Parasitol. 2010 40:109-21. Copyright Elsevier 2010.
See original on MMP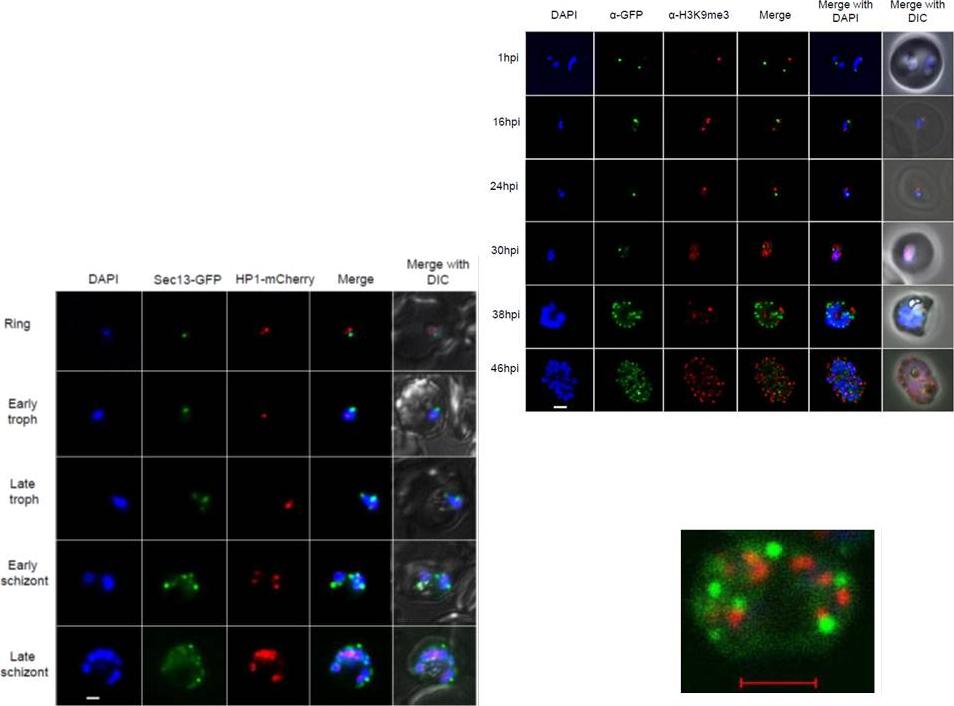
Right: PfSec13 does not co-localize with P. falciparum heterochromatin. IFAs of the PfSec13-GFP expressing parasites that were co-immunolabled with α-GFP (green) and α-H3K9me3 (red) at different stages of the IDC. In 85% of the counted nuclei of all stages (n=100), the H3K9me3 signal and PfSec13-GFP did not co-localize.Left: In vivo imaging of parasites expressing both PfSec13-GFP and PfHP1-HA-mCherry during the IDC. Although PfSec13 (green) and PfHP1 (red) localized to the nuclear periphery they do not co-localize.Righet lower corner: In vivo confocal microscopy of parasites expressing both PfSec13-GFP and PfHP1-HA-mCherry.Dahan-Pasternak N, Nasereddin A, Kolevzon N, Pe'er M, Wong W, Shinder V, Turnbull L, Whitchurch CB, Elbaum M, Gilberger TW, Yavin E, Baum J, Dzikowski R. PfSec13 is an unusual chromatin associated nucleoporin of Plasmodium falciparum, which is essential for parasite proliferation in human erythrocytes. J Cell Sci. 2013 126(Pt 14):3055-69 Reproduced with permission
See original on MMP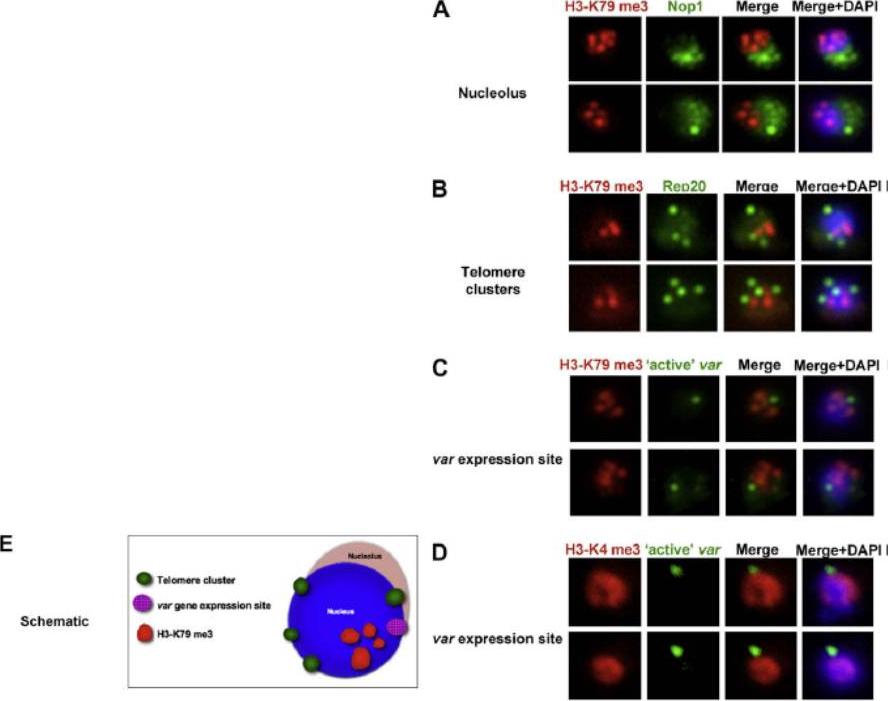
Co-localisation of H3-K79me3 with known cellular compartments. A. Nucleolus: Double labelling IFAs with antibodies against H3-K79me3 and nucleolus marker, PfNop1. B. Telomere clusters: IFeFISH analysis between H3-K79me3 (red) and Rep20 (green). C. Active var gene: IF-FISH analysis between H3-K79me3 (red) and active var gene (green). D. Active var gene colocalisation with activating mark, H3-K4me3. IF-FISH analysis between H3-K4me3 (red) and active var gene (green). DAPI stained nucleus in blue. Font colour represents corresponding fluorescein labelled antibody. Scale bar depicts 1 mm. E. Schematic of the dynamic compartmen-talisation of the P. falciparum nucleus. The distinct sub-nuclear distribution of histone methylation modification, taking example of H3-K79me3 as observed in this study. Known compartments compatible with active transcription are indicated. H3-K79me3 occupies a distinct zone.Issar N, Ralph SA, Mancio-Silva L, Keeling C, Scherf A. Differential sub-nuclear localisation of repressive and activating histone methyl modifications in P. falciparum. Microbes Infect. 2009 11:403-7.
See original on MMP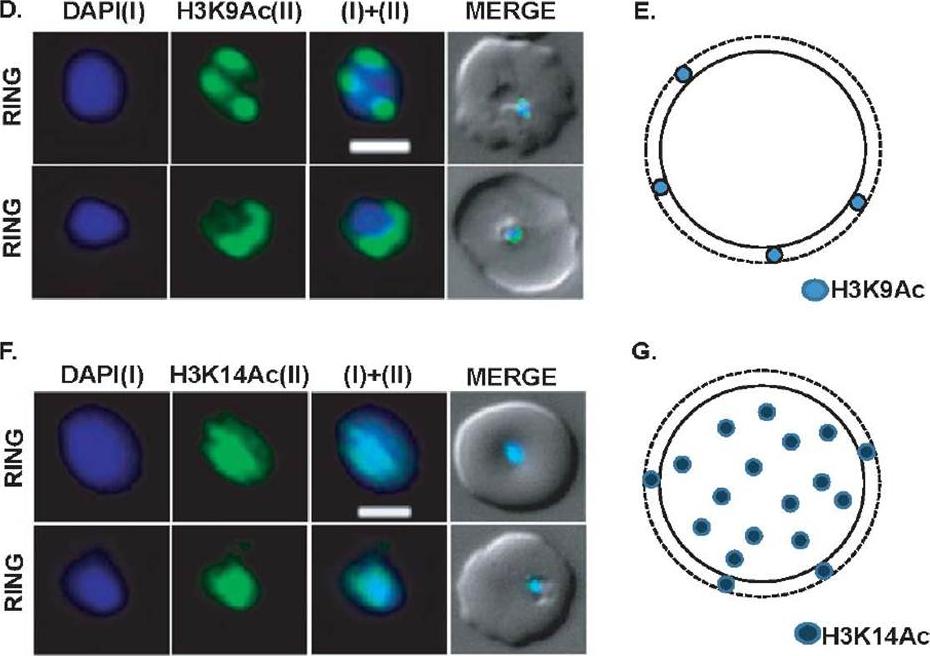
Expression and sub-cellular localization patterns of H3K9Ac and H3K14Ac histone marks. (D) Perinuclear localization pattern of acetylated H3K9Ac marks. Immunofluorescence analysis using antibodies specific for H3K9Ac confirms horseshoe-shaped pattern around nuclear periphery. H3K9Ac also localizes as sharp 3-7 perinuclear foci in majority of the parasites. (E) Schematic representation of H3K9Ac mark in parasite nuclei. (F) Localization pattern of H3K14Ac mark in the parasite nuclei. Unlike H3K9Ac, H3K14Ac signal shows even distribution on the DAPI. (G) Schematic diagram of H3K14Ac marks in the parasite nuclei. Scale bar measures 1 μM. DAPI stains ring nuclei.Srivastava S, Bhowmick K, Chatterjee S, Basha J, Kundu TK, Dhar SK. Histone H3K9 acetylation level modulates gene expression and may affect parasite growth in human malaria parasite Plasmodium falciparum. FEBS J. 2014 Sep 22. [Epub ahead of print]
See original on MMP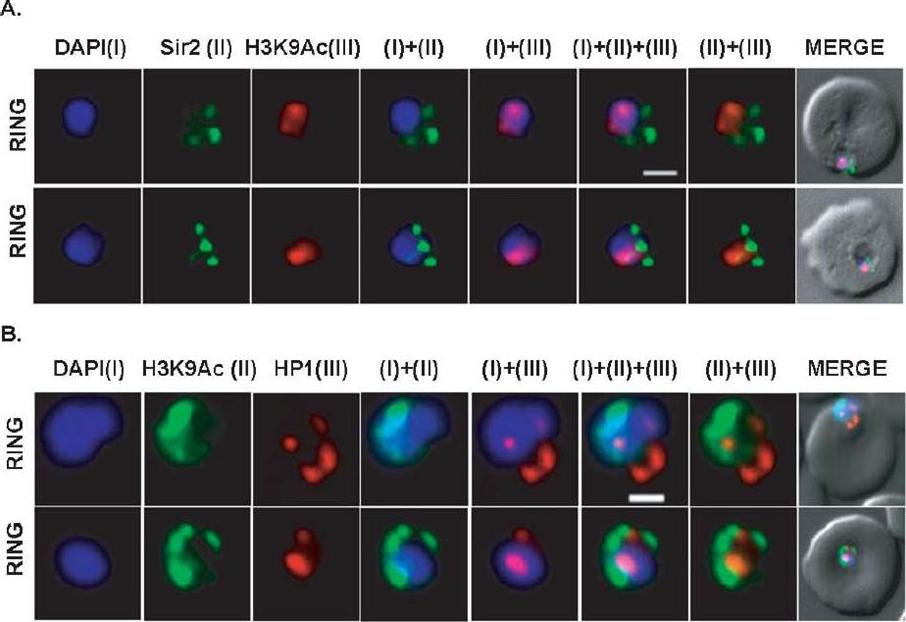
Differential localization patterns of H3K9Ac, PfSir2 and PfHP1. (A) Differential distribution of PfSir2 and H3K9Ac in Plasmodium falciparum. Dual Immuno-fluorescence analysis of PfSir2 (green) and H3K9Ac (red) reveals PfSir2 as sharp foci around nuclei and H3K9Ac as sharp foci closely attached to nuclei periphery. There is no colocalization of PfSir2 and H3K9Ac in ring stage. Scale bar measures 1 μM. DAPI stains nuclei. (B) Differential distribution of H3K9Ac (green) and PfHP1, specific for H3K9me3 repressive histone marks (red). Dual immuno-fluorescence assay shows no colocalization between these two proteins. Scale bar measures 1 μM. DAPI stains nuclei.Srivastava S, Bhowmick K, Chatterjee S, Basha J, Kundu TK, Dhar SK. Histone H3K9 acetylation level modulates gene expression and may affect parasite growth in human malaria parasite Plasmodium falciparum. FEBS J. 2014 Sep 22. [Epub ahead of print]
See original on MMP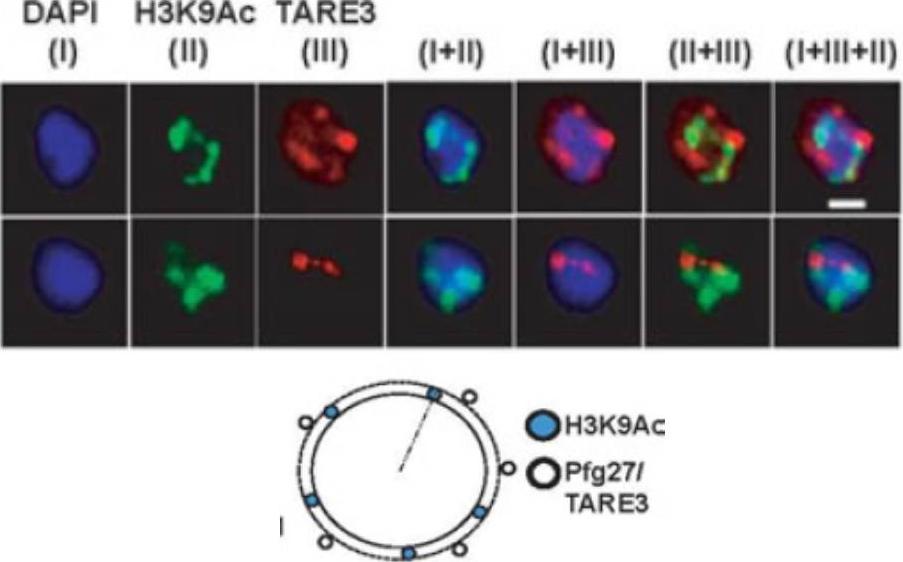
Active promoter localizes to H3K9Ac enriched zones located at nuclear periphery. No colocalization takes place between TARE3 sub-telomeric regions and H3K9Ac marks. FISH-IFA experiments were performed using labeled TARE3 (red) DNA probe and antibodies specific for H3K9Ac (green) marks. Left scheme: Schematic representation of differential localizations of TARE3 and H3K9Ac marks. DAPI stains ring nuclei. Scale bar measures 1 μM.Srivastava S, Bhowmick K, Chatterjee S, Basha J, Kundu TK, Dhar SK. Histone H3K9 acetylation level modulates gene expression and may affect parasite growth in human malaria parasite Plasmodium falciparum. FEBS J. 2014 Sep 22. [Epub ahead of print]
See original on MMPMore information
| PlasmoDB | PVP01_1221000 |
| GeneDB | PVP01_1221000 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | null |
| Orthologs | PBANKA_1348400 , PCHAS_1353000 , PF3D7_1333700 , PKNH_1266900 , PVX_082615 , PY17X_1353400 |
| Google Scholar | Search for all mentions of this gene |