PVP01_1016700 serine/arginine-rich splicing factor 1, putative (SR1)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
RMgm-5089 | Imported from RMgmDB | |
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. falciparum 3D7 | Asexual |
Possible |
34291805 (Knock down)
\"We show that a highly conserved RNA splicing factor, P. falciparum (Pf)SR1, plays an unexpected and crucial role in DNA repair in malaria parasites. Using an inducible and reversible system to manipulate PfSR1 expression, we demonstrate that this protein is recruited to foci of DNA damage. Although loss of PfSR1 does not impair parasite viability, the protein is essential for their recovery from DNA-damaging agents or exposure to artemisinin, the first-line antimalarial drug, demonstrating its necessity for DNA repair.\" |
Theo Sanderson, Francis Crick Institute |
Imaging data (from Malaria Metabolic Pathways)
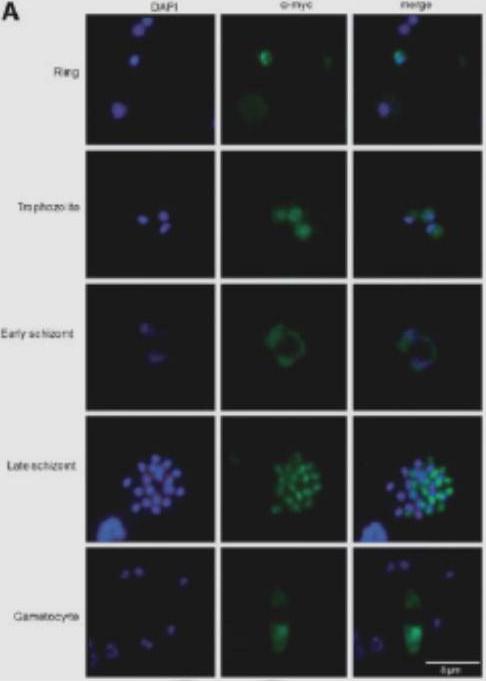
The RS domain localizes PfSR1 to the nucleus. IFAs showing the localization of PfSR1 wasectopically express fused to a myc epitope tag and detected using anti-myc antibodies. PfSR1 (green); DAPI staining (blue); developmental stages are indicated on the left. (A) Cellular localization of PfSR1-myc. PfSR1 shuttles between the nucleus and cytoplasm. The cytoplasmic location is restricted to regions that are outside the parasite’s food vacuole. the nuclear location of PfSR1 at early ring stages is in a distinct punctuated pattern mainly at the nuclear periphery and is not equally spread all over the nucleus.Eshar S, Allemand E, Sebag A, Glaser F, Muchardt C, Mandel-Gutfreund Y, Karni R, Dzikowski R. A novel Plasmodium falciparum SR protein is an alternative splicing factor required for the parasites' proliferation in human erythrocytes. Nucleic Acids Res. 2012 40(19):9903-16
See original on MMP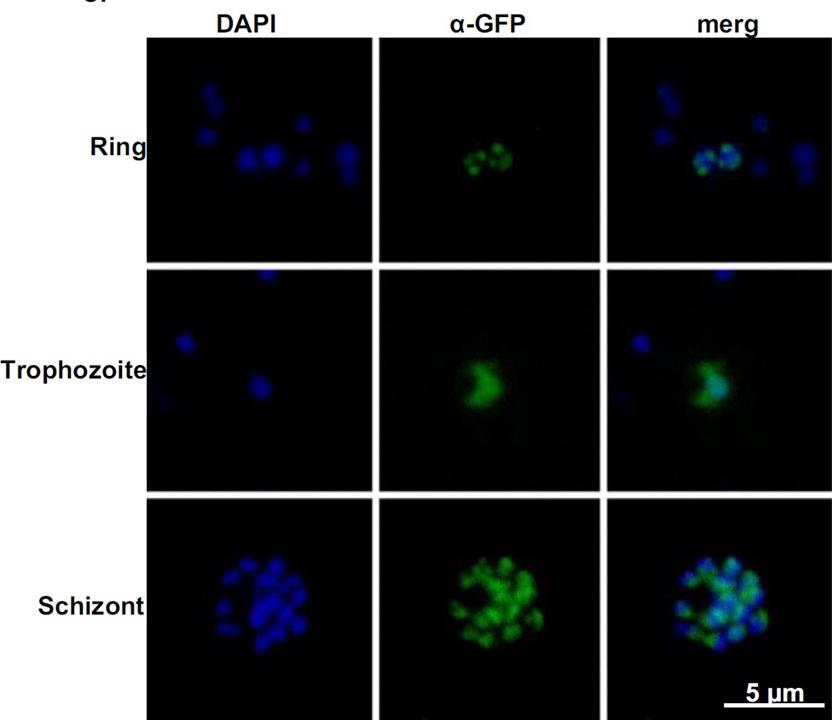
Induction of PfSR1 expression using the DDD system. Immuno-fluorescence assay (IFA) of PfSR1-GFP-DDD expressing parasites growing on 5 μM TMP showing the cellular localization of the induced PfSR1-GFP throughout the IDC. Tagged PfSR1 was detected using anti-GFP antibody. PfSR1 is shown in green; nuclei were stained with DAPI (blue). in early ring stages PfSR1-GFP-DDD localizes to the nucleus in foci that are mainly located at the nuclear periphery. This pattern changes later in the IDC at the trophozoite and schizont stages, when PfSR1-GFP-DDD can be detected in both the nucleus and in the cytoplasm .Eshar S, Altenhofen L, Rabner A, Ross P, Fastman Y, Mandel-Gutfreund Y, Karni R, Llinás M, Dzikowski R. PfSR1 controls alternative splicing and steady-state RNA levels in Plasmodium falciparum through preferential recognition of specific RNA motifs. Mol Microbiol. 2015 Jun;96(6):1283-97. a
See original on MMPMore information
| PlasmoDB | PVP01_1016700 |
| GeneDB | PVP01_1016700 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | null |
| Orthologs | PBANKA_1232100 , PCHAS_1232700 , PF3D7_0517300 , PKNH_1016000 , PVX_080385 , PY17X_1235500 |
| Google Scholar | Search for all mentions of this gene |