PVP01_0916400 peptidyl-prolyl cis-trans isomerase 11, putative
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Possible |
RMgm-253 | Imported from RMgmDB | |
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. berghei ANKA | Asexual |
No difference |
RMgm-253 | Imported from RMgmDB |
| P. berghei ANKA | Gametocyte |
No difference |
RMgm-253 | Imported from RMgmDB |
| P. berghei ANKA | Ookinete |
No difference |
RMgm-253 | Imported from RMgmDB |
| P. berghei ANKA | Oocyst |
No difference |
RMgm-253 | Imported from RMgmDB |
| P. berghei ANKA | Sporozoite |
No difference |
RMgm-253 | Imported from RMgmDB |
| P. berghei ANKA | Liver |
No difference |
RMgm-253 | Imported from RMgmDB |
Imaging data (from Malaria Metabolic Pathways)
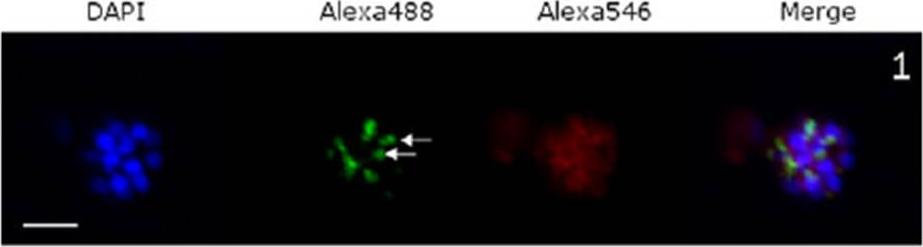
Confocal immunofluorescence microscopic images of P. falciparum schizonts treated with: Schizonts were stained with DAPI (nuclear stain), Alexafluor-488 (PfRAP1) and Alexafluor-546 (PfCYP19B). White scale bars indicate 5 μM. White arrows show characteristic bi-punctate rhoptry staining. Immuno-staining schizonts for RAP1 under normalconditions resulted in the bi-punctate staining characteristic of rhoptry proteins, while PfCYP19B waslocated in the cytoplasm as expected.Leneghan D, Bell A. Immunophilin-protein interactions in Plasmodium falciparum. Parasitology. 2015 Jul 9:1-11. [Epub ahead of print] PubMed PMID: 26156578
See original on MMP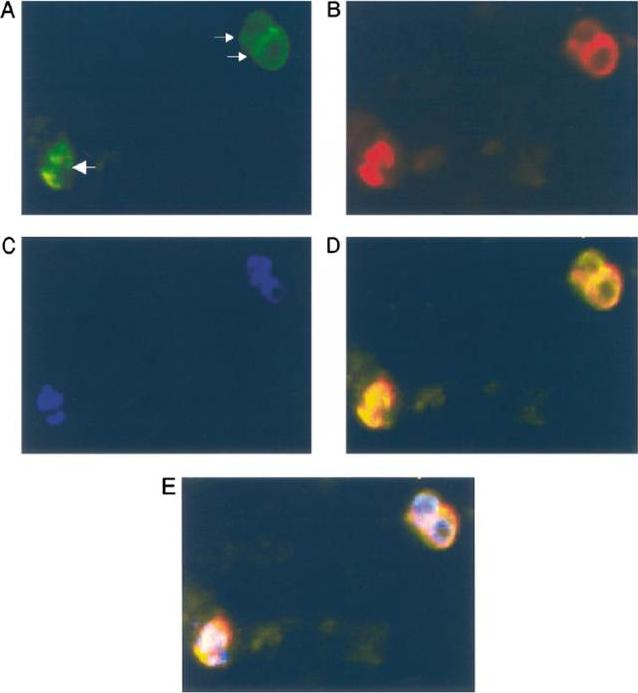
Subcellular location of PfCyP-19B determined by immunofluorescence. Trophozoite cultures were prepared for immunofluoresence as described in Section 2, using primary rabbit antibodies to PfCyP-19B and mouse antibodies to LDH, secondary FITC-conjugated anti-rabbit IgG and TRITC-conjugated anti-mouse IgG antibodies, and DAPI stain. Two singly infected erythrocytes (small arrows) and a doubly infected erythrocyte (large arrow) can be seen in this representative picture. Panel A: FITC image (PfCyP-19B); Panel B: TRITC image (LDH); Panel C: DAPI image (nuclear stain); Panel D: FITC and TRITC images superimposed; Panel E: FITC, TRITC, and DAPI images superimposed. Experiments using the different fluorophores separately confirmed that there was no significant cross-fluorescence using any of the filter sets. the protein appeared surprisingly to be distributed throughout the cell, as though it were located in the cytosol.Gavigan CS, Kiely SP, Hirtzlin J, Bell A. Cyclosporin-binding proteins of Plasmodium falciparum. Int J Parasitol. 2003 Aug;33(9):987-96.
See original on MMPMore information
| PlasmoDB | PVP01_0916400 |
| GeneDB | PVP01_0916400 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | null |
| Orthologs | PBANKA_0932200 , PCHAS_0912100 , PF3D7_1115600 , PKNH_0913200 , PVX_091425 , PY17X_0934200 |
| Google Scholar | Search for all mentions of this gene |