PVP01_0819700 nicotinamidase, putative (Nico)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Possible |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. berghei ANKA | Asexual |
No difference |
PlasmoGEM (Barseq) | PlasmoGEM |
Imaging data (from Malaria Metabolic Pathways)
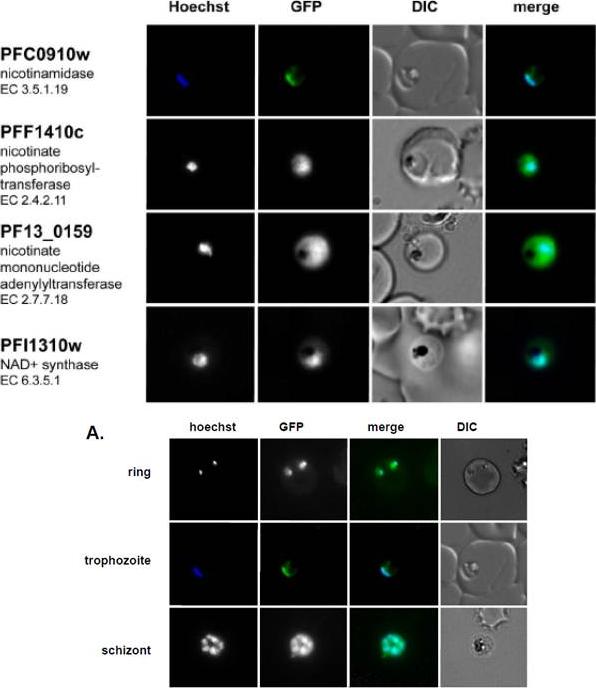
Live imaging of episomally expressed GFP tagged NAD+ metabolic enzymes are shown (GFP-fusion proteins are shown in green). Enzyme Commission numbers are provided for each enzyme. All images are of trophozoite stage parasites. Hoechst dye (shown in blue) was used to visualize the parasite nucleus. NAD+ pathway enzymes are primarily localized in the cytoplasm except for the parasite nicotinamidase, which localizes to the cytoplasm but concentrates mainly in the nucleus.Live imaging of the episomally expressed PfNico-GFP fusion. The nucleus is visualized with Hoechst staining. O'Hara JK, Kerwin LJ, Cobbold SA, Tai J, Bedell TA, Reider PJ, Llinás M. Targeting NAD+ Metabolism in the Human Malaria Parasite Plasmodium falciparum. PLoS One. 2014 Apr 18;9(4):e94061.
See original on MMPMore information
| PlasmoDB | PVP01_0819700 |
| GeneDB | PVP01_0819700 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | null |
| Orthologs | PBANKA_1218000 , PCHAS_1218700 , PF3D7_0320500 , PKNH_0820300 , PVX_095210 , PY17X_1221200 |
| Google Scholar | Search for all mentions of this gene |