PVP01_0422600 early transcribed membrane protein (ETRAMP11.2)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
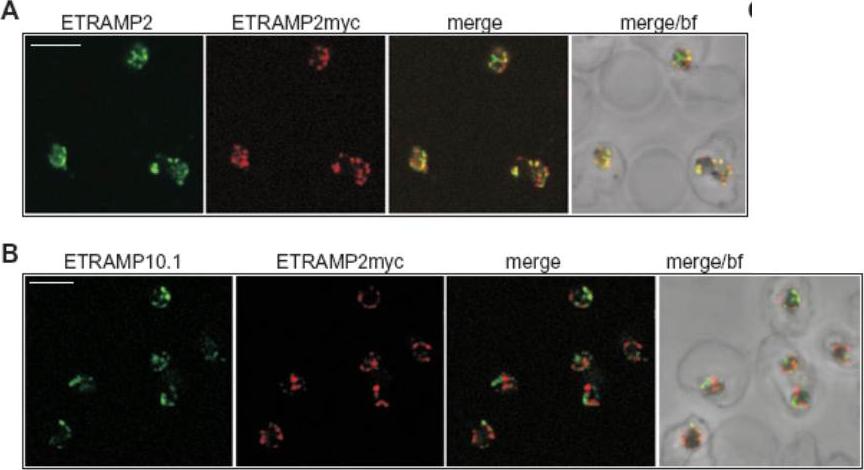
Confocal microscopy demonstrates patching of different ETRAMPs into distinct domains in formaldehyde-fixed IRBCs. IRBCs (D10E2myc parasites) coated on 10-well slides using concanavalinA (fixed with 1% formaldehyde and permeabilized with 0.03% saponin) were incubated with sera specific for ETRAMP2, ETRAMP10.1, ETRAMP2myc (detected via the tag) or anti-ETRAMP2 serum labelled with a fluorescent Fab fragment and viewed by confocal microscopy. A. ETRAMP2 (cy2, green) co-patches with ETRAMP2myc (cy3, red) into distinct areas in the PVM of several ring-stage IRBCs. B. ETRAMP10.1 (cy2, green) patches into distinct areas when compared with ETRAMP2myc (cy3, red) in several ring-stage IRBCs. bf, brightfield; bar size, 4 μm.Spielmann T, Gardiner DL, Beck HP, Trenholme KR, Kemp DJ. Organization of ETRAMPs and EXP-1 at the parasite-host cell interface of malaria parasites. Mol Microbiol. 2006 59:779-94. PMID:
See original on MMP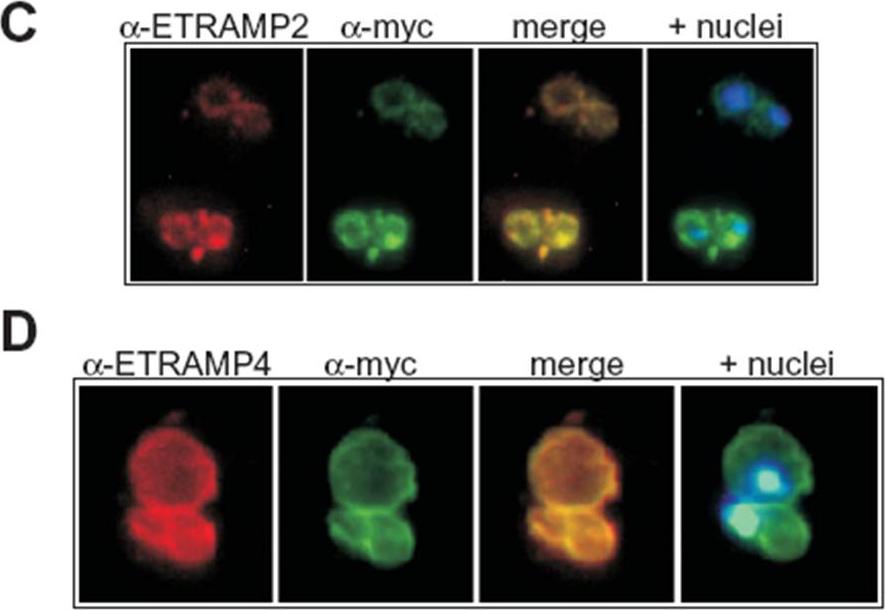
Transgenic expression of myc-tagged ETRAMP2 and ETRAMP4. Immunofluorescence analysis in acetone-fixed IRBCs: the tagged transgenes detected via the myc tag colocalize with the respective chromosomally derived ETRAMPs in a typical peripheral staining in the transfected parasites D10E2myc (C) and D10E4myc (D). C shows two IRBCs, each with a double infection of ring-stage parasites and D shows a double infection of trophozoites. Hoechst was used to stain the nuclei. Immunofluorescence analysis with an anti-myc serum showed a circular staining pattern in acetone-fixed D10E2myc and D10E4myc parasites (C and D), which is typical of the ETRAMP staining pattern seen with anti-ETRAMP serum in untransfected parasites. ETRAMPs are seen in focused areas of the PVM.Spielmann T, Gardiner DL, Beck HP, Trenholme KR, Kemp DJ. Organization of ETRAMPs and EXP-1 at the parasite-host cell interface of malaria parasites. Mol Microbiol. 2006 59:779-94.
See original on MMP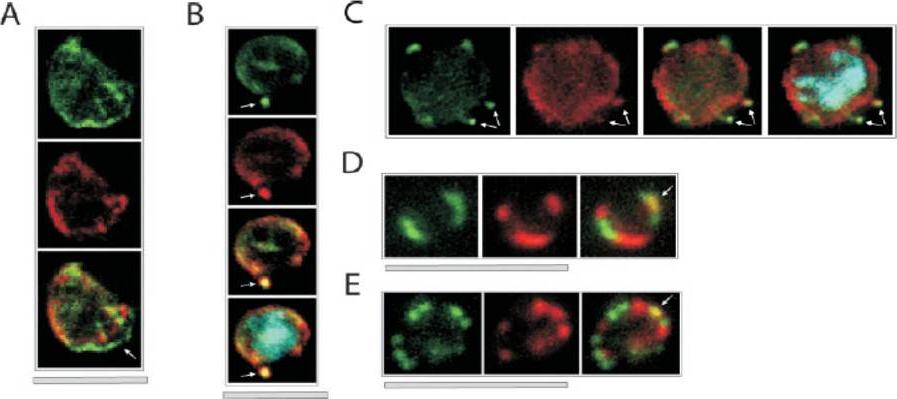
Colocalization of ETRAMPs with EXP-1 and SERP. The first image in each set shows ETRAMP labeling (green, FITC), the second Texas-Red-labeled SERP (A) or EXP-1 (B–F), and the third image represents the merged picture. The fourth panel in B and C depicts the merged picture, including nuclear staining (DAPI). (A) Confocal microscopy analysis of SERP and ETRAMP4 colocalization in schizont stage parasites (dried and 1% formaldehyde fixed). The ETRAMP4 signal seems partially detached from the PV content (arrow). (B and C) ETRAMP4 and EXP-1 localize to dots, probably representing vesicular structures that are detached from the PVM (E) or seem connected to the PVM (F). Dots are indicated by arrows. (D and E) Fluorescent microscopic analysis of ETRAMP and EXP-1 costaining of a ring-stage parasite displaying an even circular pattern (D, 0.1% formaldehyde-fixed IRBCs, ETRAMP10.1) or a beads on a string pattern (E, unfixed IRBCs, ETRAMP2). Yellow color shows regions of overlap (arrows). This indicated that ETRAMPs and EXP-1 localize to different regions of the PVM.Spielmann T, Fergusen DJ, Beck HP. etramps, a new Plasmodium falciparum gene family coding for developmentally regulated and highly charged membrane proteins located at the parasite-host cell interface. Mol Biol Cell. 2003 14:1529-44.
See original on MMP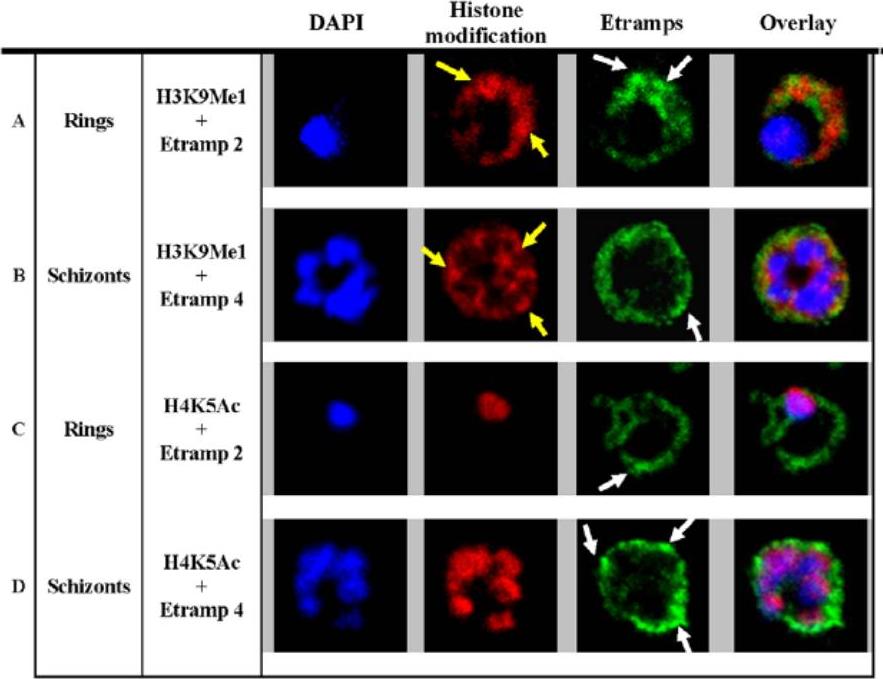
H3K9Me1 localized to the parasitophorous vacuole during the ring stage. Co-localization of H3K9Me1 with Etramp 2 (A) and 4 (B) was performed in ring and schizont stage parasites respectively. Similarly, co-localization of H4K5Ac with Etramp 2 (C) and 4 (D) was performed in ring and schizont stage parasites respectively. H3K9Me1/H4K5Ac and Etramp 2/4 were stained red and green respectively. DAPI stained nuclear DNA blue. Yellow and white arrows indicate foci of more intense fluorescence produced by H3K9Me1 and Etramp labeling respectively. In ring stage parasites, compared to schizonts, H3K9Me1 partially co-localized with Etramp 2 indicating localization to different compartments of the PV. H4K5Ac was localized solely to the nucleus and did not co-localize with either Etramp 2 or 4.Luah YH, Chaal BK, Ong EZ, Bozdech Z. A moonlighting function of Plasmodium falciparum histone 3, mono-methylated at lysine 9? PLoS One. 2010 5(4):e10252.
See original on MMPMore information
| PlasmoDB | PVP01_0422600 |
| GeneDB | PVP01_0422600 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | null |
| Orthologs | PF3D7_0202500 , PKNH_0418600 , PVX_003565 |
| Google Scholar | Search for all mentions of this gene |