PKNH_1271600 transcriptional regulatory protein sir2 homologue, putative (SIR2A)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Possible |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. berghei ANKA |
Possible |
RMgm-4435 | Imported from RMgmDB | |
| P. falciparum 3D7 |
Possible |
19402747 Longer telomeric repeats, var gene derepression. |
Theo Sanderson, Wellcome Trust Sanger Institute | |
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
| P. falciparum 3D7 |
Possible |
23443558 \"Using a parasite line in which PfSir2a has been disrupted, we observe that histones near the transcription start sites of all ribosomal RNA genes are hyperacetylated and that transcription of ribosomal RNA genes is upregulated.\" |
Theo Sanderson, Francis Crick Institute | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. berghei ANKA | Asexual |
No difference |
PlasmoGEM (Barseq) | PlasmoGEM |
| P. berghei ANKA | Asexual |
No difference |
RMgm-4435 | Imported from RMgmDB |
| P. falciparum 3D7 | Asexual |
Transcriptional effect |
23443558 \"Using a parasite line in which PfSir2a has been disrupted, we observe that histones near the transcription start sites of all ribosomal RNA genes are hyperacetylated and that transcription of ribosomal RNA genes is upregulated.\" |
Theo Sanderson, Francis Crick Institute |
| P. berghei ANKA | Gametocyte |
No difference |
RMgm-4435 | Imported from RMgmDB |
| P. berghei ANKA | Ookinete |
Difference from wild-type |
RMgm-4435
Normal gamete production, fertilisation and ookinete production. Normal formation of mature ookinetes at the light microscope level. Evidence is found that ookinetes do not traverse the the midgut epithelium. |
Imported from RMgmDB |
| P. berghei ANKA | Oocyst |
Difference from wild-type |
RMgm-4435
No oocyst formation |
Imported from RMgmDB |
| P. berghei ANKA | Sporozoite |
Difference from wild-type |
RMgm-4435
No oocyst formation. No sporozoite formation |
Imported from RMgmDB |
Imaging data (from Malaria Metabolic Pathways)
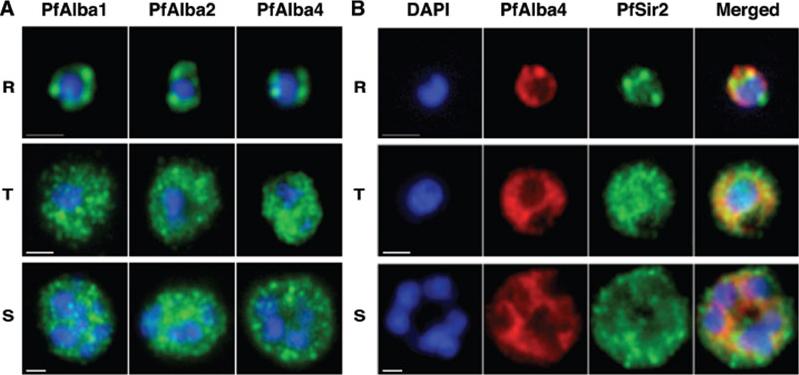
PfAlbas localize to perinuclear foci in ring stages and expand to the cytoplasm in mature forms. (A) Immunofluorescence analysis of PfAlba1, PfAlba2 and Pfalba4 distribution throughout the parasite life cycle (Rings, Trophozoites, Schizonts). The anti-PfAlba antibodies (Ab 74–2 for PfAlba1; Ab 233 for PfAlba2; Ab 237–2 for PfAlba4) were used at 5.5, 4.5 and 8 mg/ml, respectively. Anti-rabbit Alexa-488 secondary antibodies (Molecular Probes) were used at a dilution of 1:500. Scale bars represent 1 mm. (B) Immunofluorescence analysis of 3D7 Rings, Trophozoites and Schizonts stained with anti-PfAlba4 (red) and anti-PfSir2 (green) antibodies. Scale bar represents 1 mm.Chêne A, Vembar SS, Rivière L, Lopez-Rubio JJ, Claes A, Siegel TN, Sakamoto H, Scheidig-Benatar C, Hernandez-Rivas R, Scherf A. PfAlbas constitute a new eukaryotic DNA/RNA-binding protein family in malaria parasites. Nucleic Acids Res. 2012 40(7):3066-77
See original on MMP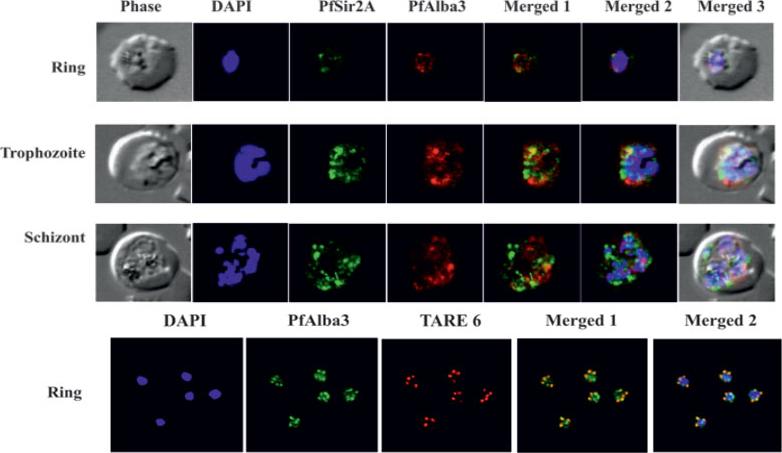
Upper panels: Plasmodium falciparum infected red blood cells were treated with DAPI (blue), mouse anti-PfSir2A antibody (secondary antibody Alexa Fluor 488, green) and rabbit anti-PfAlba3 antibody (secondary antibody Alexa Fluor 633, red), respectively. Overlay panels show the merged images in different combinations. PfAlba3 (red) and PfSir2A (green) fluorescence signals merge at several places, foremost at the peripheral region of the nucleus. However, co-localization was partial and not significant during schizont stage. Lower panel: FISH analysis combined with IF labeling of PfAlba3 to visualize telomeric region of chromosomes. Immunolocalization (IF) of PfAlba3 was performed at ring stage parasites using an Alexa 488-conjugated secondary antibody (green). FISH analysis was performed simultaneously. The nuclei were stained with DAPI (blue) and hybridized with TARE6 (Red) probe to visualize the chromosome ends on ring stage. Goyal M, Alam A, Iqbal MS, Dey S, Bindu S, Pal C, Banerjee A, Chakrabarti S, Bandyopadhyay U. Identification and molecular characterization of an Alba-family protein from human malaria parasite Plasmodium falciparum. Nucleic Acids Res. 2011 40(3):1174-90.
See original on MMP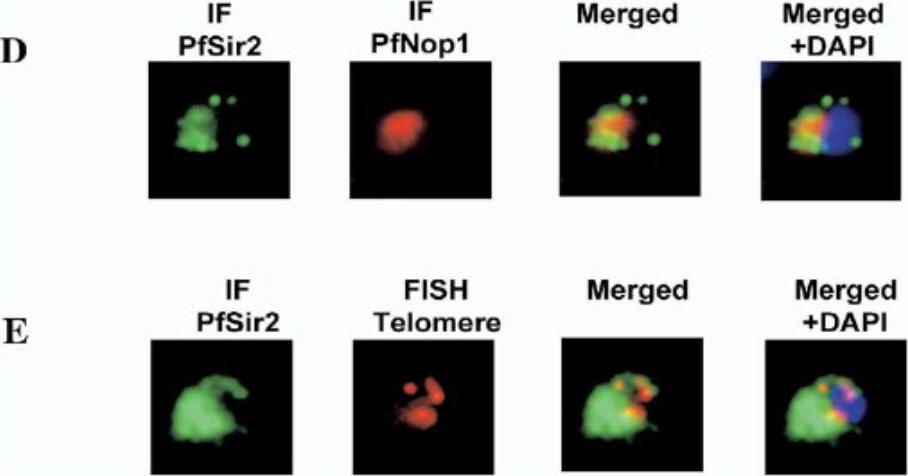
(D) IF using the PfNop1 and PfSir2 antibodies. The PfSir2 antibody is visualized in red and the PfNop1 antibody in green using Alexa 488. (E) Simultaneous IF and FISH analysis. Immunolocalization (IF) of PfSir2 was performed on young trophozoite parasites using an Alexa 488-conjugated secondary antibody (green). FISH analysis was performed simultaneously using a telomeric oligonucleotide probe conjugated with Cy3 (red). The nuclei were stained with DAPI. Protein localizes to telomeric foci. anti-PfSir2 antibody signal is highly concentrated at one pole of the nucleus.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mâncio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005 121:25-36. Copyright Elsevier 2009.
See original on MMP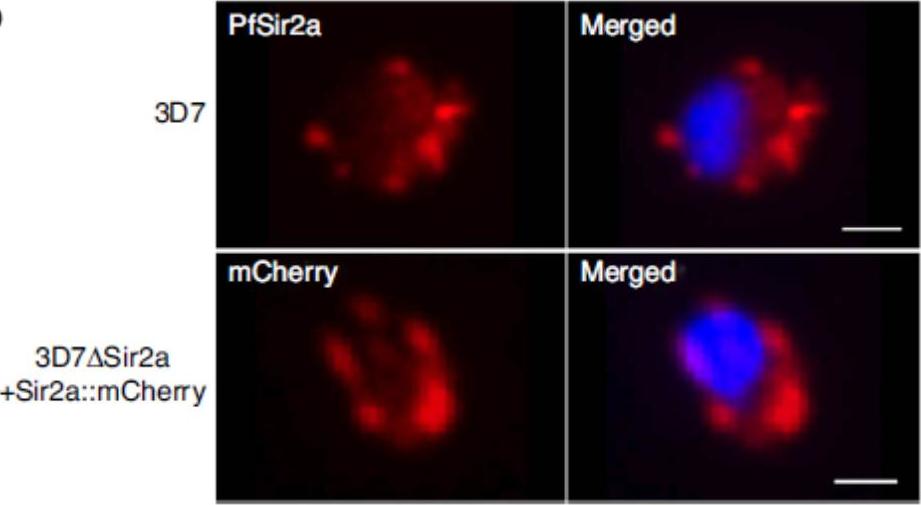
Immunofluorescence assay in paraformaldehyde fixed ring-stage parasites with anti-PfSir2a antibodies (red) in wild-type parasites (first row) and anti-mCherry antibodies (red) in 3D7DSir2aþSir2aHmCherry parasites (second row). Nuclei of ring-stage parasites were stained with DAPI (blue). Scale bar, 1 mm. PfSir2aHmCherry was enriched at the nuclear periphery, forming foci that resemble the perinuclear pattern (telomeric clusters and nucleolus) previously shown for the native PfSir2a.Mancio-Silva L, Lopez-Rubio JJ, Claes A, Scherf A. Sir2a regulates rDNA transcription and multiplication rate in the human malaria parasite Plasmodium falciparum. Nat Commun. 2013 4:1530.
See original on MMP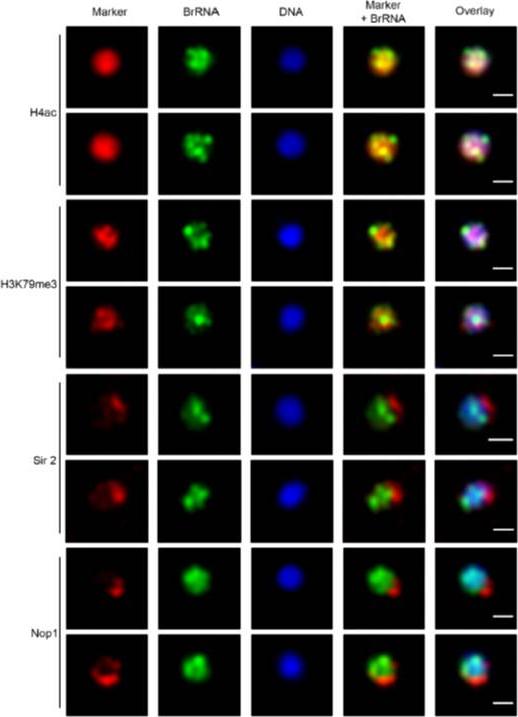
Transcription sites form a distinct compartment in the nucleus of P. falciparum. Immunofluorescence assay for nascent RNAs of ring stage parasites labeled with BrUTP (BrRNA, green) and the compartments of acetylated H4 (H4ac, active chromatin); a proposed histone marker of nascent transcription (H3K79me3; PfSir2A (silencing compartment); and Nop1 (nucleolar compartment). DNA was labeled by DAPI, in blue. Bars, 1 mm.Moraes CB, Dorval T, Contreras-Dominguez M, Dossin Fde M, Hansen MA, Genovesio A, Freitas-Junior LH. Transcription sites are developmentally regulated during the asexual cycle of Plasmodium falciparum. PLoS One. 2013;8(2):e55539.
See original on MMP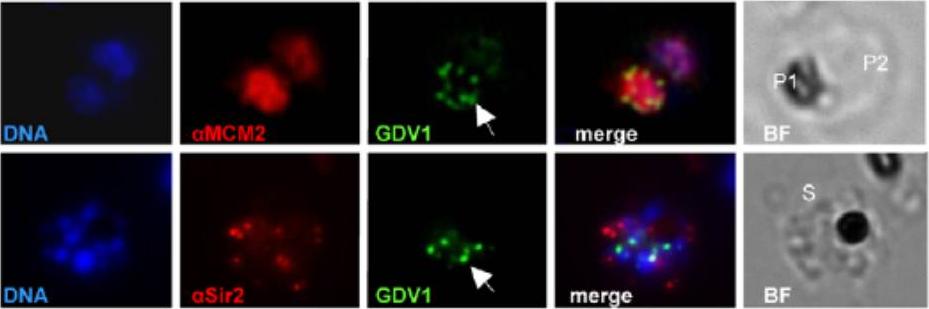
Subcellular localization of PfGDV1 (this gene product is expressed in gametocytes and is not included in MPMP). Parasites transformed with GFP- PfGDV1 were stained with DAPI (DNA stain) and the indicated anti-sera, and then examined by fluorescence microscopy (Zeiss Axiovert 200, 1000xmagnification). Images are shown of the DAPI stain (DNA), GFP-tagged PfGDV1 epifluorescence (GDV1), and antibodies specific for the indicated proteins. The corresponding merged and bright field (BF) images are included on the right. PfGDV1 expression is indicated with an arrow; locations of parasites in the BF image are indicated with a P for parasite or S for schizont. Colocalization of PfGDV1 with nuclear proteins. A doubly infected erythrocyte (Upper) with one parasite in the plane of the image (P1) and the other below (P2). Both P1 and P2 are positive for GDV1 and aMCM2. A schizont (S) expressing GDV1 stained with aSir2. MCM2 localized to the nucleus, and PfSir2 to the nuclear periphery.Eksi S, Morahan BJ, Haile Y, Furuya T, Jiang H, Ali O, Xu H, Kiattibutr K, Suri A, Czesny B, Adeyemo A, Myers TG, Sattabongkot J, Su XZ, Williamson KC. Plasmodium falciparum gametocyte development 1 (Pfgdv1) and gametocytogenesis early gene identification and commitment to sexual development. PLoS Pathog. 2012 8(10):e1002964.
See original on MMP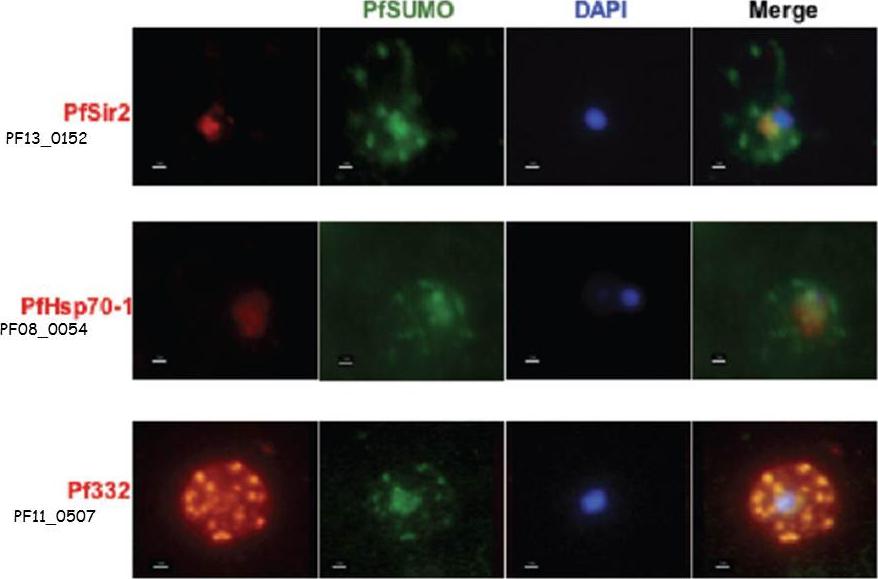
Co-localization assays of PfSUMO antiserum with nuclear marker PfSir2, cytoplasmic marker, PfHsp70-1 and Maurer’s clefts marker, Pf332 reveal compartmentalization. Size bar corresponds to 1 mm.Issar N, Roux E, Mattei D, Scherf A. Identification of a novel post-translational modification in Plasmodium falciparum: protein sumoylation in different cellular compartments. Cell Microbiol. 2008 10:1999-2011.
See original on MMP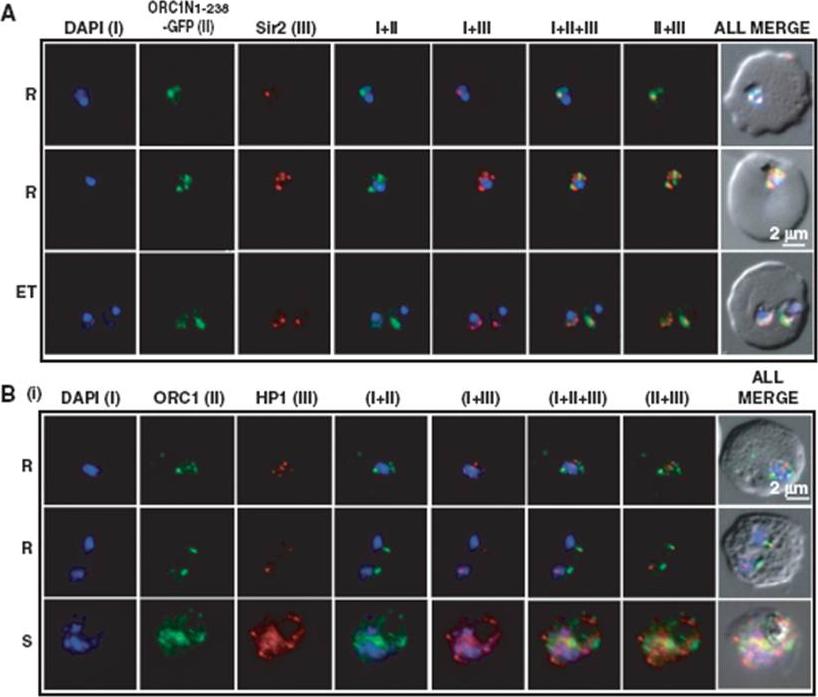
Co-localization of ORC1N1–238–GFP and Sir2 and immunolocalization of ORC1 and HP1 in 3D7. Glass slides containing the parasite smears from ORC1N1–238–GFP expressing parasites or 3D7 wild-type parasite lines were treated for immunolocalization using respective antibodies as indicated in the panels (A) and (B). (A) Co-localization of GFP and Sir2 in ORC1N1–238–GFP parasites during early stages of development as indicated on the left. ‘R’ indicates ring stage and ‘ET’ indicates early trophozoite stage parasites. DAPI shows the nuclei. Both ORC1 and Sir2 are localized to the nuclear periphery during the ring or early trophozoite stage. However, both proteins reorganize themselves at the onset of DNA replication and spread in the nucleus and cytoplasm. B(i): Both ORC1 and HP1 show nuclear punctate staining in 3D7 wild-type parasites during the early stages of development. ‘S’ indicates schizont stage parasites.Deshmukh AS, Srivastava S, Herrmann S, Gupta A, Mitra P, Gilberger TW, Dhar SK. The role of N-terminus of Plasmodium falciparum ORC1 in telomeric localization and var gene silencing. Nucleic Acids Res. 2012 40(12):5313-31
See original on MMP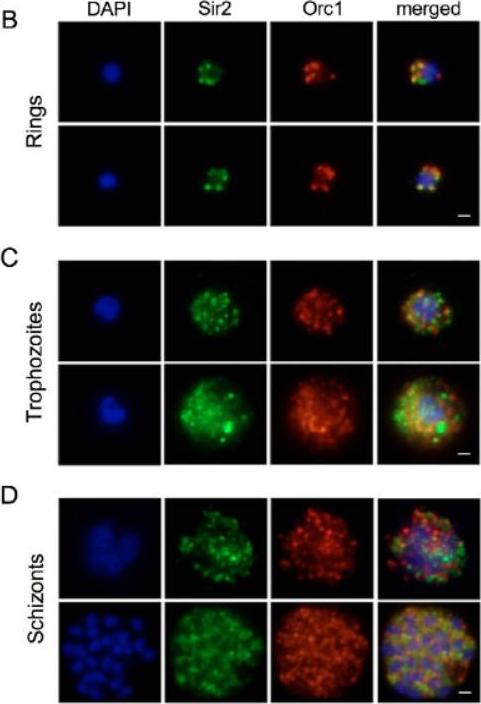
Sir2 and Orc1 relocalize during the developmental cycle of P. falciparum. (B-D) IF analysis of Sir2 (green) and Orc1 (red) during the blood-stage cycle. (B) Ring stages display a punctate pattern at the nuclear periphery. (C) In the trophozoite stages, anti-Sir2 and -Orc1 antibodies reveal an apparent increase of both protein levels, and an additional punctate and diffuse pattern inside and outside of the nucleus. (D) The schizont stage, Sir2 and Orc1 seem to relocalize at the nuclear periphery.Mancio-Silva L, Rojas-Meza AP, Vargas M, Scherf A, Hernandez-Rivas R. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum. J Cell Sci. 2008 121:2046-53.
See original on MMP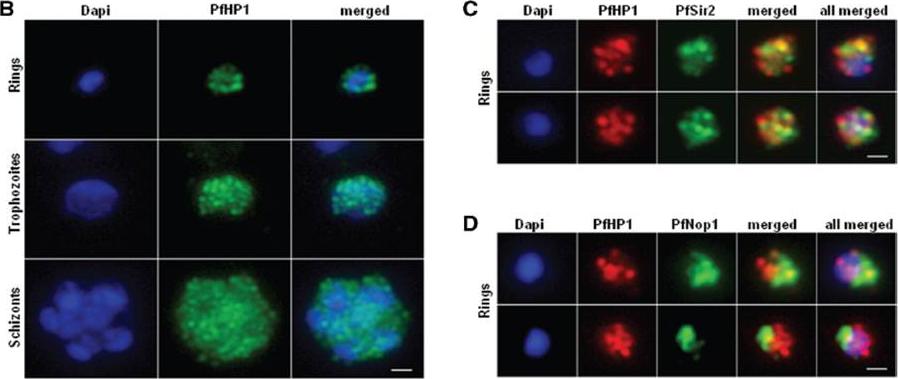
Nuclear expression and cellular localization of PfHP1. (B) IFA analysis of PfHP1 (green) throughout the parasite asexual blood stage cycle (ring, trophozoite and schizont stages). (C) In ring stage, partial co-localization between PfHP1 (red) and PfSir2 (green) antibodies (yellow spots) is observed. (D) Co-localization with a nucleolar marker PfNop1 shows minimal overlap of PfHP1 with this compartment. Scale bars represent 1 mm.Pérez-Toledo K, Rojas-Meza AP, Mancio-Silva L, Hernández-Cuevas NA, Delgadillo DM, Vargas M, Martínez-Calvillo S, Scherf A, Hernandez-Rivas R. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009 37(8):2596-606
See original on MMP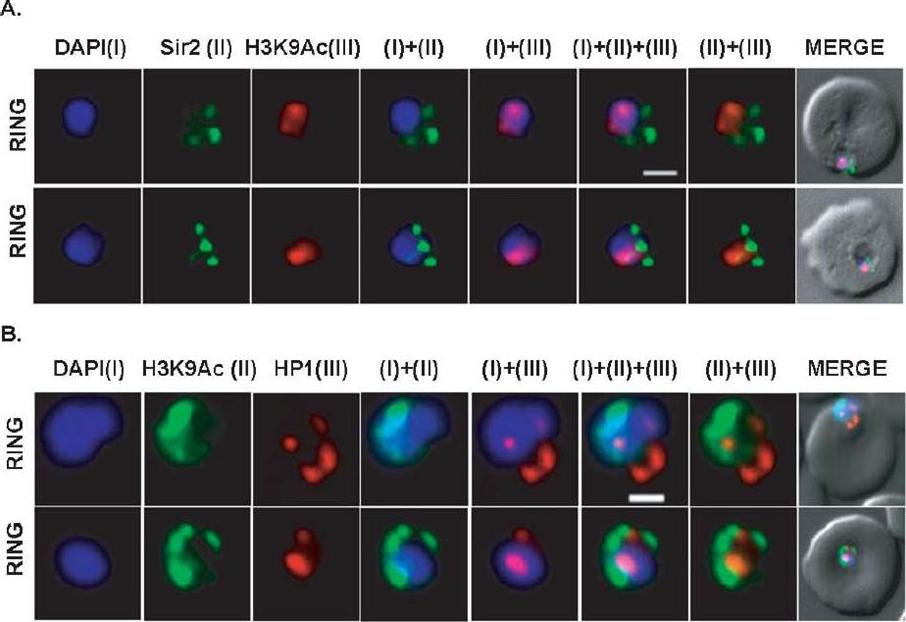
Differential localization patterns of H3K9Ac, PfSir2 and PfHP1. (A) Differential distribution of PfSir2 and H3K9Ac in Plasmodium falciparum. Dual Immuno-fluorescence analysis of PfSir2 (green) and H3K9Ac (red) reveals PfSir2 as sharp foci around nuclei and H3K9Ac as sharp foci closely attached to nuclei periphery. There is no colocalization of PfSir2 and H3K9Ac in ring stage. Scale bar measures 1 μM. DAPI stains nuclei. (B) Differential distribution of H3K9Ac (green) and PfHP1, specific for H3K9me3 repressive histone marks (red). Dual immuno-fluorescence assay shows no colocalization between these two proteins. Scale bar measures 1 μM. DAPI stains nuclei.Srivastava S, Bhowmick K, Chatterjee S, Basha J, Kundu TK, Dhar SK. Histone H3K9 acetylation level modulates gene expression and may affect parasite growth in human malaria parasite Plasmodium falciparum. FEBS J. 2014 Sep 22. [Epub ahead of print]
See original on MMPMore information
| PlasmoDB | PKNH_1271600 |
| GeneDB | PKNH_1271600 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PK13_2930w, PKH_122390 |
| Orthologs | PBANKA_1343800 , PCHAS_1348400 , PF3D7_1328800 , PVP01_1225700 , PVX_082385 , PY17X_1348600 |
| Google Scholar | Search for all mentions of this gene |