PKNH_1138500 conserved Plasmodium protein, unknown function
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
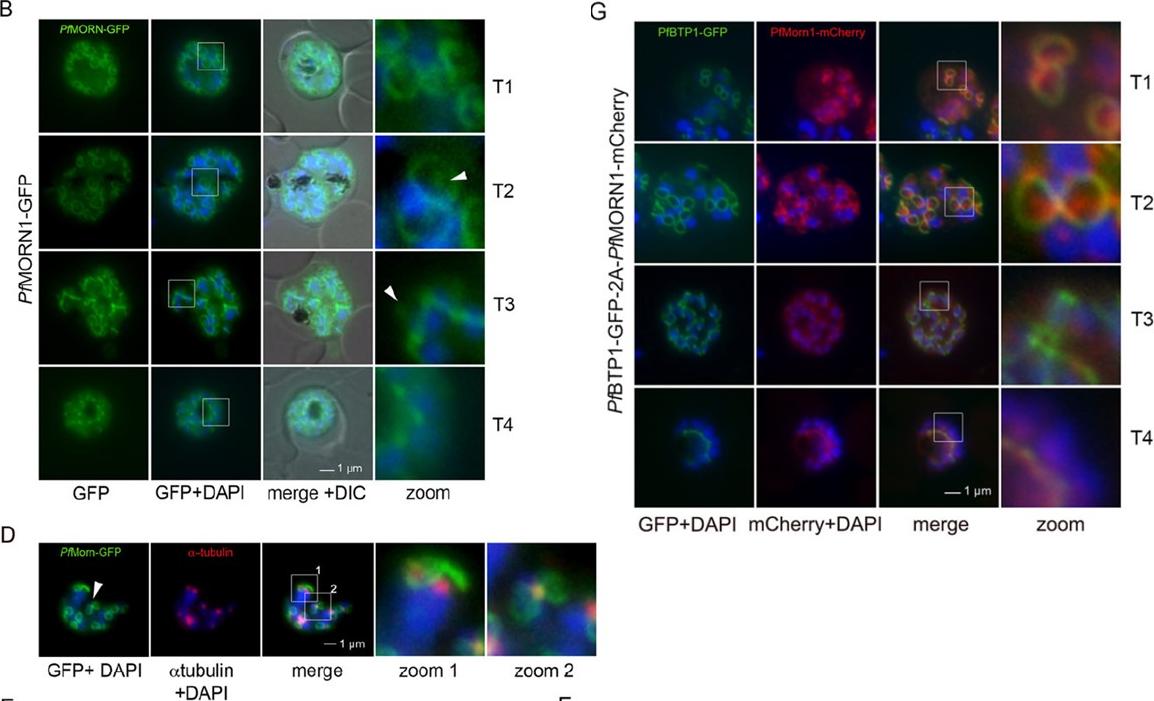
Colocalization of BTP1 with MORN1. (B) Localization of P. falciparum (pf)MORN1–GFP in unfixed parasites. MORN1–GFP reveals a contractile ring structure that moves along the longitudinal axes of the nascent merozoite (T1–T4) with an additional punctate structure that might represent the centrocone (marked with arrowheads). Nuclei were stained with DAPI (blue). Enlargement of selected areas are marked with a white square and referred to as ‘zoom’ (a fourfold magnification). (D) Colocalization of MORN1–GFP with α-tubulin (red) that highlights the microtubule-organizing centers (MTOCs) at this stage. Although the MORN1-marked ring structure is clearly distinct from the MTOC (zoom 1), there is a close association of the MTOC with the additional MORN1-marked structure (zoom 2). (G) Colocalization analysis of BTP1–GFP (green) with MORN1–mCherry (red) revealed their identical localization during schizogony, except for the exclusively MORN1–mCherry-highlighted putative centrocone.Kono M, Heincke D, Wilcke L, Wong TW, Bruns C, Herrmann S, Spielmann T, Gilberger TW. Pellicle formation in the malaria parasite. J Cell Sci. 2016 129(4):673-80. PMID: 26763910
See original on MMP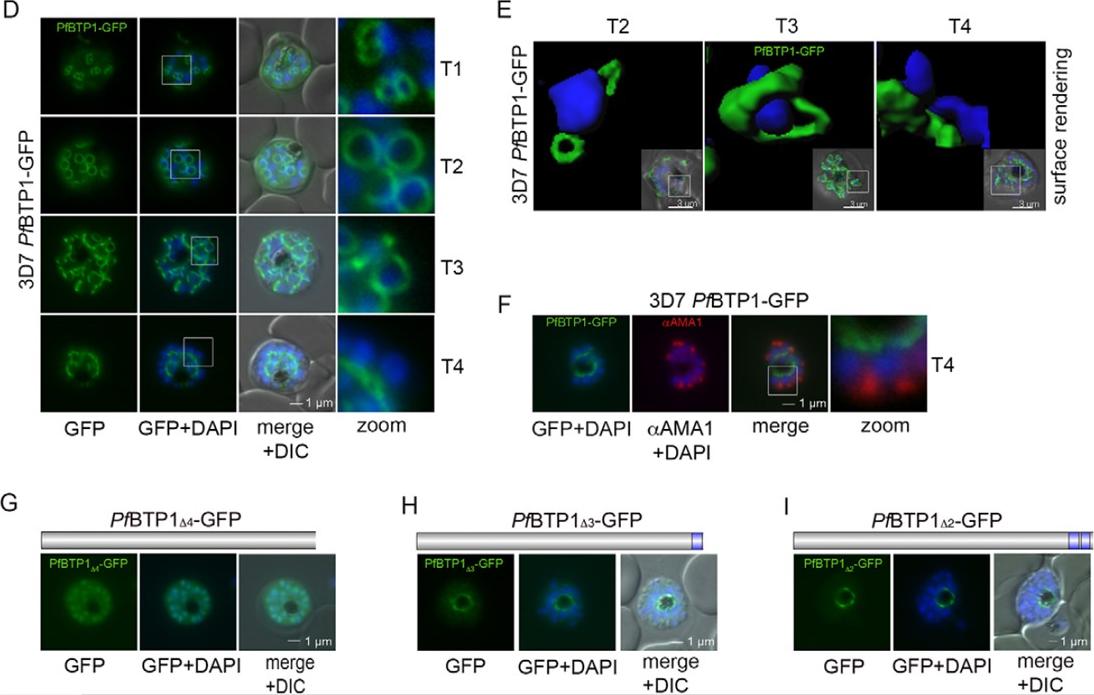
Expression and localization of BTP1. (D) BTP1–GFP can be visualized as a contractile ring structure in unfixed late-stage parasites (T1–T4). It first appears at the apical pole of the nascent merozoite (T1) and ends up at the basal pole of the mature merozoite (T4). Nuclei were stained with DAPI (blue). Enlargement of selected areas are marked with a white square and referred to as ‘zoom’ (a fourfold magnification). (E) Surface-rendering plot of an indicated section, highlighting the spatial relationship between nuclei and the basal-complex ring. (F) Colocalization of BTP1–GFP and the apical membrane antigen 1 (αAMA1; red); AMA1 localizes in the micronomes at the apical pole opposing BPT1, confirming the basal localization of BTP1–GFP at the end of daughter cell formation. (G–I) Localization of the BPT1 deletion mutants. (G) Deletion of all four transmembrane domains leads to a nuclear and cytosolic distribution of BTP1Δ4−GFP. (H) Expression of BTP1 with one (BTP1Δ3−GFP) or two (BTP1Δ2−GFP, I) transmembrane domains results in localization of mutant BTP1 at the food vacuole membrane. DIC, differential interference contrast.Kono M, Heincke D, Wilcke L, Wong TW, Bruns C, Herrmann S, Spielmann T, Gilberger TW. Pellicle formation in the malaria parasite. J Cell Sci. 2016 129(4):673-80.
See original on MMP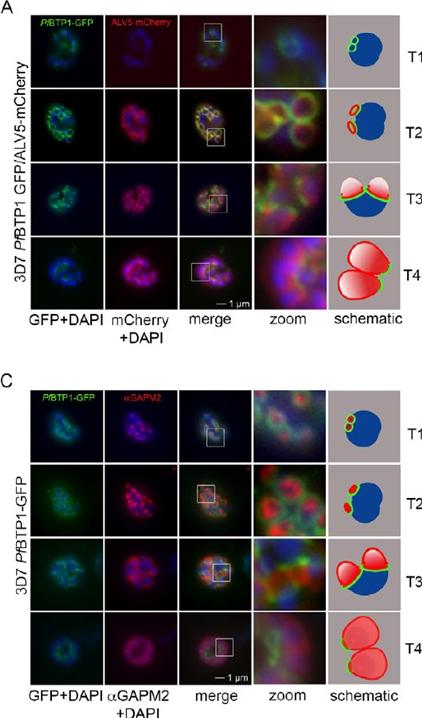
Colocalization of BTP1 with ALV5 and GAPM2 during inner membrane complex (IMC) biogenesis. (A) Colocalization of BTP1–GFP with ALV5–mCherry (red) in unfixed parasites. The initial ring structure marked by BTP1–GFP precedes the appearance of Alveolin 5 (ALV5) (T1). The emerging Alveolin-5-defined ring is associated but distinct from that marked by BTP1 (T2). During IMC expansion, BTP1 remains rim associated (T3) and, at the end of daughter cell formation, is concentrated at the basal end with ALV5 equally distributed within the IMC (T4). (C) Colocalization of BTP1–GFP with GAPM2 (red) in fixed parasites. Both proteins appeared at about the same time, marking the onset of IMC biogenesis (T1). GAPM2 stains small ring–shaped formations that are encircled by BTP1–GFP (T2). With elongation and propagation of the IMC from the apical towards the basal end of the forming daughter cell, GAPM2 is equally distributed within the forming IMC, but BTP1 is only present at the rim of the nascent IMC (T3) and, at the end of schizogony, is concentrated at the basal pole of the daughter cell (T4). Nuclei were stained with DAPI (blue). Enlargement of selected areas are marked with a white square and referred to as ‘zoom’ (a fourfold magnification). The bottom row shows a schematic representation of the spatio-temporal distribution of BTP1 and GAPM2. Kono M, Heincke D, Wilcke L, Wong TW, Bruns C, Herrmann S, Spielmann T, Gilberger TW. Pellicle formation in the malaria parasite. J Cell Sci. 2016 129(4):673-80.
See original on MMP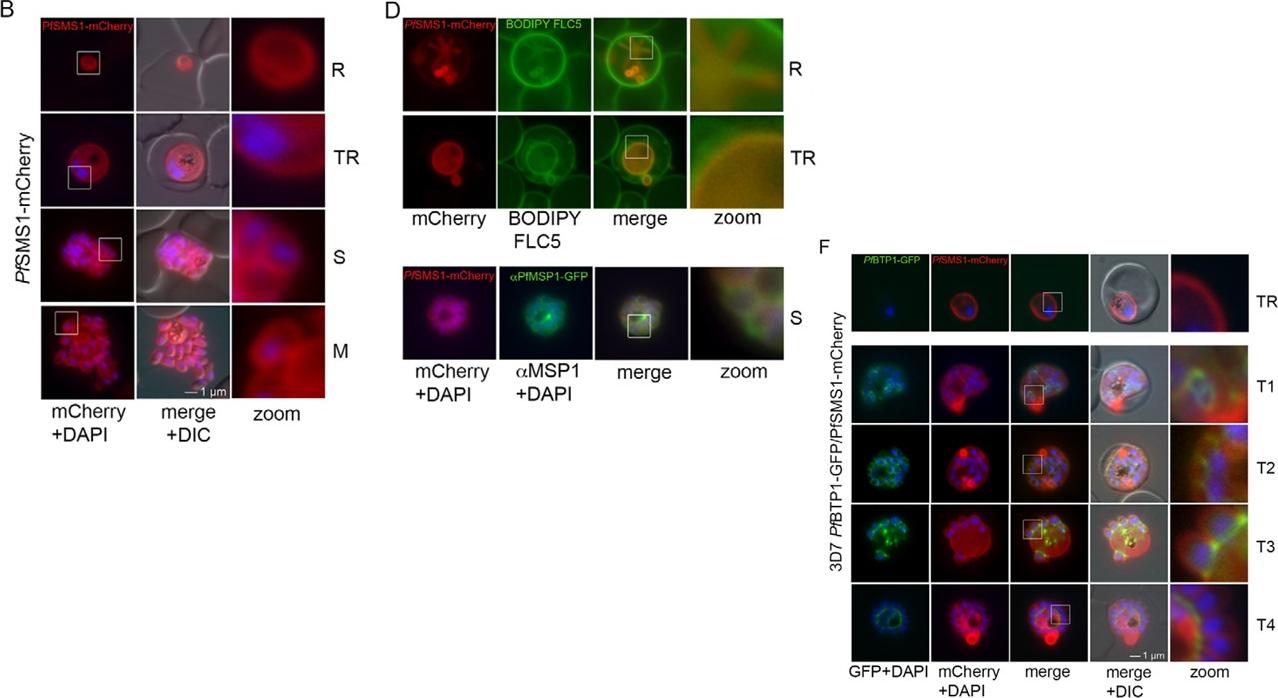
Parasite plasma membrane dynamics and correlation with BTP1. (A) Schematic of the structure of SMS1 (PFF1210w). Predicted transmembranedomains are indicated in blue. (B) Localization of SMS1–mCherry (red) during asexual proliferation in rings (R), trophozoites (TR), schizonts (S) and merozoites(M). Nuclei were stained with DAPI (blue). Enlargement of selected areas are marked with a white square and referred to as ‘zoom’ (a fourfold magnification).Please also refer to Fig. 3 and Movie 3. (C) Expression of SMS1–mCherry was analyzed by western blotting analysis using antibodies against mCherry(α-mCherry), resulting in the detection of a fusion protein of approximately 80 kDa (calculated molecular mass 78 kDa). (D) Colocalization of SMS1–mCherry withthe membrane marker fluorophore-labeled BODIPY©-FL C5-sphingomyelin lipid (upper panels) or the plasma membrane surface protein MSP1 (lower panel;αMSP1). (F) Colocalization of BTP1–GFP (green) with SMS1–mCherry (red) in trophozoites (TR) and during schizogony (T1–T4). Nuclei were stained with DAPI (blue). Enlargement of selected areas are marked with a white square and referred to as ‘zoom’. (G) Quantification of the colocalization analysis of BTP1–GFP with SMS1–mCherry (please refer to F). (H) Expression of BTP1–GFP and SMS1–mCherry was confirmed by western blotting analysis using antibodies against GFP and mCherry. DIC, differential interference contrastKono M, Heincke D, Wilcke L, Wong TW, Bruns C, Herrmann S, Spielmann T, Gilberger TW. Pellicle formation in the malaria parasite. J Cell Sci. 2016 129(4):673-80.
See original on MMPMore information
| PlasmoDB | PKNH_1138500 |
| GeneDB | PKNH_1138500 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PK6_0990c, PKH_113720 |
| Orthologs | PBANKA_0109900 , PCHAS_0110500 , PF3D7_0611600 , PVP01_1137600 , PVX_113725 , PY17X_0111500 |
| Google Scholar | Search for all mentions of this gene |