PF3D7_0501200 parasite-infected erythrocyte surface protein (PIESP2)
Disruptability [+]
| Species | Disruptability | Reference | Submitter |
|---|---|---|---|
| P. falciparum 3D7 |
Possible |
18614010 | Theo Sanderson, Wellcome Trust Sanger Institute |
| P. falciparum 3D7 |
Possible |
29100811 Some trafficking defects and altered Maurers Clefts |
Theo Sanderson, Wellcome Trust Sanger Institute |
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
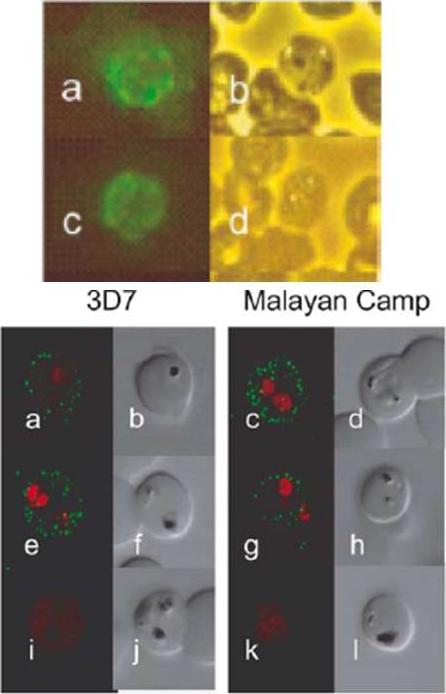
Localization of PIESP1 and PIESP2 with specific antibodies. (A) Immunofluorescent microscopy of PIEs (P. falciparum strain 3D7) fixed onto glass slides. Glass slide were incubated with anti-PIESP1 (Plates a and b), anti-PIESP2 (Plates c and d), and pooled preimmune sera (Plates e and f). Fluorescent images (Plates a, c, and e) and phase-contrast images (Plates b, d, and f) were collected simultaneously on an Olympus BX-60. (B) Localization of PIESPs with immunofluorescent confocal microscopy. Live P. falciparum strains 3D7 and Malayan Camp (unselected for knob and rosetting phenotypes) were incubated in suspension with specified antibodies. Plates a–d, anti-PIESP1 as primary antibody; Plates e–h, anti-PIESP2 as primary antibody; Plates i–l, preimmune sera as primary antibody. The 488 and 568 nm laser channels, for Alexa Fluor 488 and ethidine bromide, respectively, were collected separately and later superimposed (Plates a, c, e, g, i, and k). The DIC images (Plates b, d, f, h, j, and l) were collected simultaneously using the transmitted light detector. Florens L, Liu X, Wang Y, Yang S, Schwartz O, Peglar M, Carucci DJ, Yates JR 3rd, Wub Y. Proteomics approach reveals novel proteins on the surface of malaria-infected erythrocytes. Mol Biochem Parasitol. 2004 135:1-11. Copyright Elsevier 2010.
See original on MMP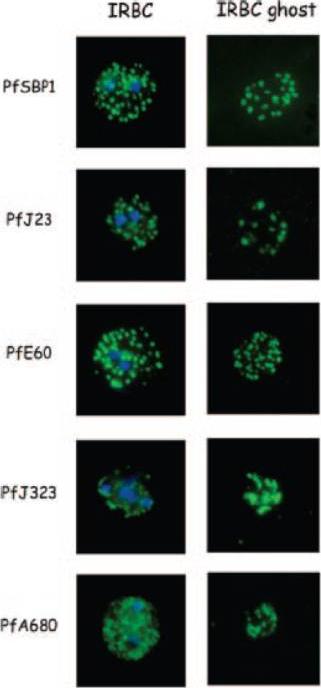
Indirect immunofluorescence of P. falciparum-infected erythrocytes and infected red blood cell (IRBC) ghosts. Air-dried infected red blood cells and infected red blood cell ghosts were incubated with mouse antibodies raised against GST fusion proteins as indicated. The nuclei were stained with DAPI (blue). Negative controls were performed using preimmune sera and anti-GST antibodies (not shown). All proteins are associated with Maurer;s clefts.Vincensini L, Richert S, Blisnick T, Van Dorsselaer A, Leize-Wagner E, Rabilloud T, Braun Breton C. Proteomic analysis identifies novel proteins of the Maurer's clefts, a secretory compartment delivering Plasmodium falciparum proteins to the surface of its host cell. Mol Cell Proteomics. 2005 4:582-93.
See original on MMP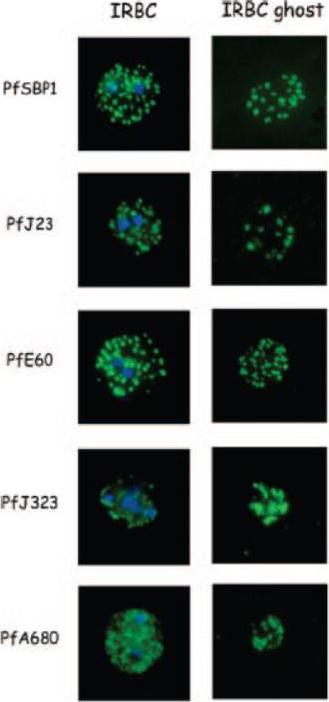
Indirect immunofluorescence of P. falciparum-infected erythrocytes and infected red blood cell (IRBC) ghosts. Air-dried infected red blood cells and infected red blood cell ghosts were incubated with mouse antibodies raised against GST fusion proteins as indicated. The nuclei were stained with DAPI (blue). Negative controls were performed using preimmune sera and anti-GST antibodies (not shown). All proteins are associated with Maurer;s clefts.Vincensini L, Richert S, Blisnick T, Van Dorsselaer A, Leize-Wagner E, Rabilloud T, Braun Breton C. Proteomic analysis identifies novel proteins of the Maurer's clefts, a secretory compartment delivering Plasmodium falciparum proteins to the surface of its host cell. Mol Cell Proteomics. 2005 4:582-93.
See original on MMP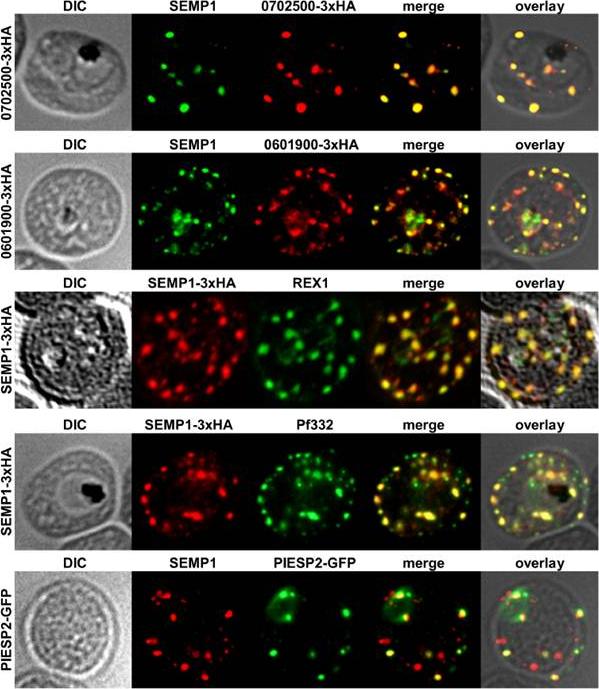
Localization of potential SEMP1 interacting proteins. Immunofluorescence assays of MeOH-fixed RBCs infected with 3D7 expressing EMP1/PF3D7_0702500 (PF07_0008)/PF3D7_0601900 (PFF0090w) with a C-terminal 3xHA tag and 3D7 expressing PIESP2 with a C-terminal GFP-tag, co-labelled with mouse a-SEMP1 and rat a-HA (PF3D7_0702500-3xHA & PF3D7_0601900-3xHA) /a-GFP (PIESP2-GFP). For co-labelling of SEMP1-3xHA with REX1 and Pf332, rat a-HA and rabbit a-REX1 / mouse a-Pf332 antibodies were used. All potential interaction partners co-localized at least partially with SEMP1, but in particular with Pf332 or PIESP2 antibodies only a fraction of SEMP1 positive MCs were labelled with either antibody suggesting that distinct subpopulations of MCs exist. Dietz O, Rusch S, Brand F, Mundwiler-Pachlatko E, Gaida A, Voss T, Beck HP. Characterization of the Small Exported Plasmodium falciparum Membrane Protein SEMP1. PLoS One. 2014 9(7):e103272.
See original on MMP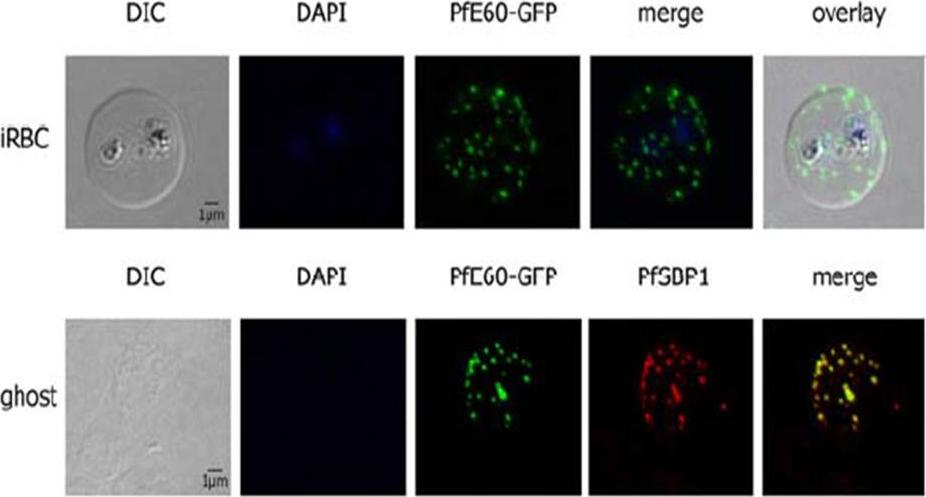
PfTRiC is interacting with the PEXEL-positive PfE60 Maurer’s cleft membrane protein in the cytoplasm of 3D7 iRBCs. Fluorescent patterns of iRBCs infected by 3D7/pARL2-PfE60-gfp (live imaging) and of resealed ghosts from 3D7/pARL2-PfE60-gfp iRBCs (GFP fluorescence, in green, and immunodetection using anti-SBP1 antibodies, in red). RBCs and ghost preparations were incubated with DAPI for nucleus labelling. The GFP-tagged PfE60 protein was detected in the membrane of Maurer’s clefts as shown previously for native PfE60.Mbengue A, Vialla E, Berry L, Fall G, Audiger N, Demettre-Verceil E, Boteller D, Braun-Breton C. New export pathway in Plasmodium falciparum-infected erythrocytes: role of the parasite group II chaperonin, PfTRiC. Traffic. 2015 Jan 23. [Epub ahead of print]
See original on MMPMore information
| PlasmoDB | PF3D7_0501200 |
| GeneDB | PF3D7_0501200 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | MAL5P1.13, PFE0060w |
| Orthologs | |
| Google Scholar | Search for all mentions of this gene |