PF3D7_0412700 erythrocyte membrane protein 1, PfEMP1 (VAR)
Disruptability [+]
| Species | Disruptability | Reference | Submitter |
|---|---|---|---|
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
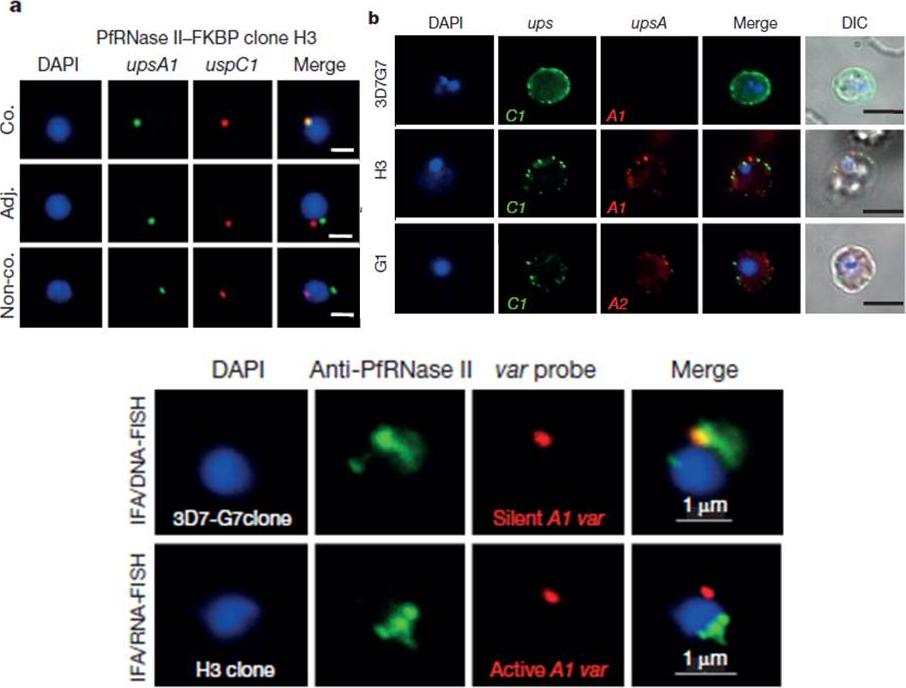
Monoallelic expression of var gene family is controlled by chromatin-associated RNase II (PFI0295c). a, Two-colour RNA-FISH of var transcripts in PfRNase II–FKBP-H3 and G1 clones. Red, UpsC-type C1 var probe (PFD0625c); green, upsA-type A1 var (PFF0020c). Co., co-localization; adj., adjacent; non-co., non-co-localization. Nuclear DNA was stained by DAPI (blue). Scale bar, 1 mm. b, Live-cell IFA using antisera to various PfEMP1s to detect co-expression of var genes in PfRNase II–FKBP H3 and G1 clones, respectively. The 3D7-G7 control expresses only one type of PfEMP1 (C1 var). Scale bar, 5 mm. c, PfRNase II is linked to the silent var-associated loci. Left: combined FISH/IFA assay using anti-PfRNase II antibody (IFA) with a FISHprobe (PFF0020c) that detected the upsA var expression site (RNA-FISH) in PfRNase II–FKBP clone H3. The same upsA var gene (DNA-FISH/IFA) was also analysed in ring-stage 3D7-G7 parasites.Zhang Q, Siegel TN, Martins RM, Wang F, Cao J, Gao Q, Cheng X, Jiang L, Hon CC, Scheidig-Benatar C, Sakamoto H, Turner L, Jensen AT, Claes A, Guizetti J, Malmquist NA, Scherf A. Exonuclease-mediated degradation of nascent RNA silences genes linked to severe malaria. Nature. 2014 Jun 29.
See original on MMPMore information
| PlasmoDB | PF3D7_0412700 |
| GeneDB | PF3D7_0412700 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | MAL4P2.58, PFD0625c |
| Orthologs | |
| Google Scholar | Search for all mentions of this gene |