PCHAS_1321400 aminopeptidase P, putative (APP)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Possible |
RMgm-813 | Imported from RMgmDB | |
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. berghei ANKA | Asexual |
Difference from wild-type |
RMgm-813
Blood stages show a strongly reduced growth rate in mice and produce strongly reduced levels of hemozoin compared to wild type parasites. |
Imported from RMgmDB |
Imaging data (from Malaria Metabolic Pathways)
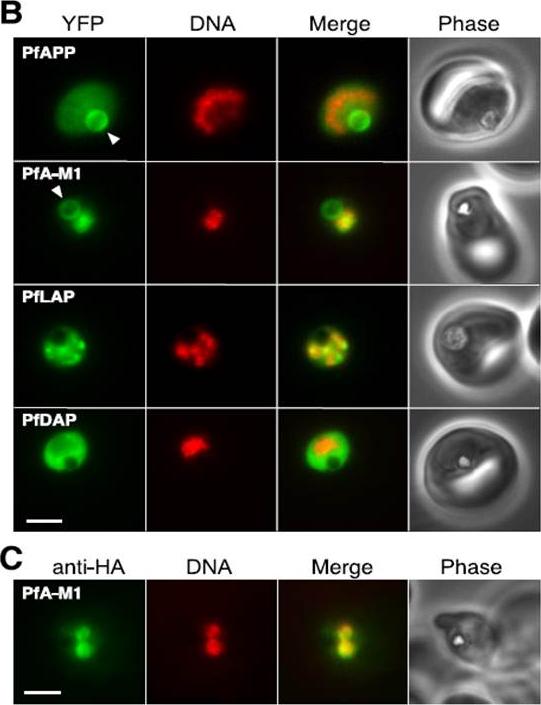
Localization of aminopeptidases in P. falciparum blood stages.PfAPP Aminopeptidase P; PfA-M1 Aminopeptidase N; PfLAP Leucyl aminopeptidase; PfDAP Aspartyl aminopeptidase B, Epifluorescence microscopy of live parasites expressing fusions of YFP with the indicated aminopeptidases. YFP fluorescence is pseudocolored green and Hoechst 33342 (DNA) fluorescence is pseudocolored red. The arrowhead indicates the food vacuole. Bar, 2 mm. C, indirect immunofluorescence detection of PfA-M1-HA. Secondary antibody fluorescence is pseudocolored green and DAPI (DNA) fluorescence is pseudocolored red. Bar, 2 mm.Dalal S, Klemba M. Roles for two aminopeptidases in vacuolar hemoglobin catabolism in Plasmodium falciparum. J Biol Chem. 2007 282:35978-87.
See original on MMP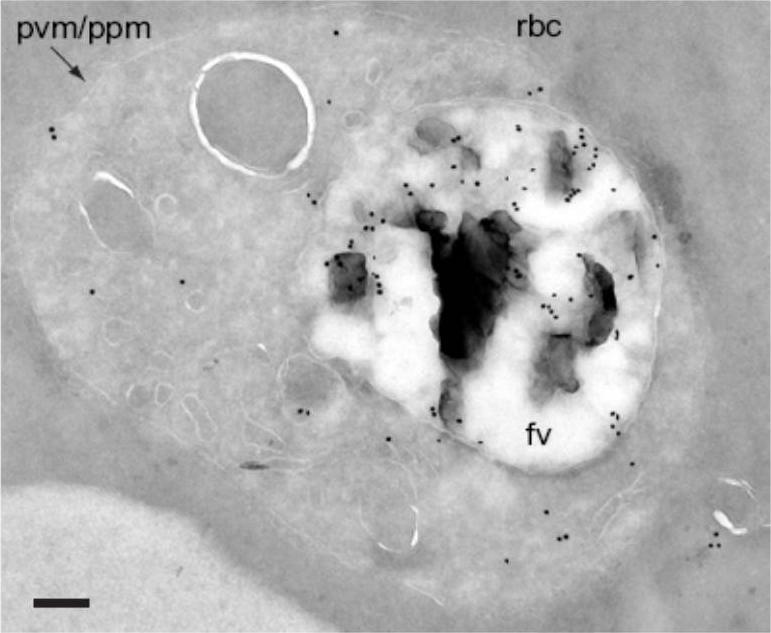
Localization of PfAPP by cryoimmunoelectron microscopy. Parasitized erythrocytes were fixed with 4% paraformaldehyde and 0.1% glutaraldehyde and were labeled with anti-PfAPP serum (1:2,500 dilution). rbc, red blood cell; fv, food vacuole; pvm/ppm, parasitophorous vacuole membrane/parasite plasma membrane. Bar, 200 nm. The distribution of gold particles was 79 % in the food vacuole and 21 % outside of the food vacuole but within the parasite.Ragheb D, Bompiani K, Dalal S, Klemba M. Evidence for catalytic roles for Plasmodium falciparum aminopeptidase P in the food vacuole and cytosol. J Biol Chem. 2009 284(37):24806-15 PubMed
See original on MMPMore information
| PlasmoDB | PCHAS_1321400 |
| GeneDB | PCHAS_1321400 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PC000641.01.0, PC000971.01.0, PC300522.00.0, PC302381.00.0, PCAS_132140, PCHAS_132140 |
| Orthologs | PBANKA_1318100 , PF3D7_1454400 , PKNH_1227700 , PVP01_1252600 , PVX_117760 , PY17X_1321900 |
| Google Scholar | Search for all mentions of this gene |