PCHAS_0935100 SET domain protein, putative
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
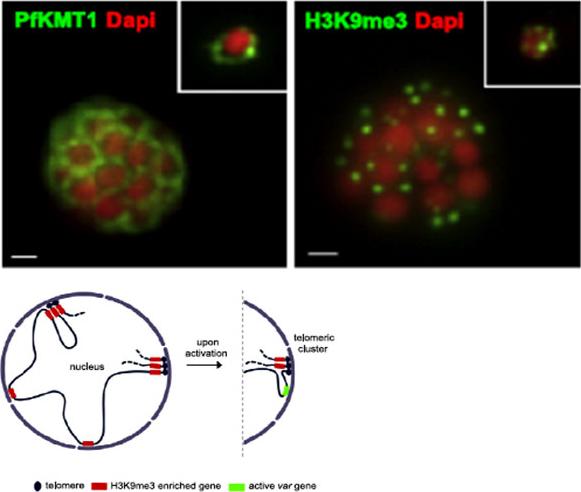
PfKMT1 and genes enriched in H3K9me3 are positioned at the nuclear periphery. Immuno-fluorescence assays using antibodies against H3K9me3 H3 (right) and the P. falciparum putative H3K9 methyltransferase PfKMT1 (left). 3D7 parasites are in late schizont (main image) and ring stage (top-right square). Nuclear DNA was detected with DAPI (red).Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009 5:179-90.
See original on MMP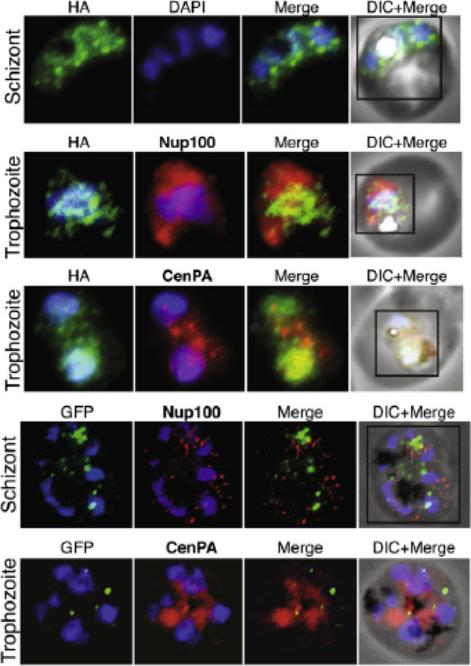
SET-domain proteins PF08_0012 and Histone-lysine N-methyltransferase PF13_0293 localize to the nuclear periphery marked by CenPA. (A) Localization of PF08_0012 reveals a clustered localization pattern present throughout the nuclear periphery of the cell. PF08_0012 is HA-tagged at the C-terminus and detected with anti-HA antibodies. (B) Live imaging of PF13_0293 shows localization to a distinct area in the nuclear periphery. Co-localization studies by immunofluorescence are shown. Nup100 (PFI0250c) and CenPA (histone H3 PF13_0185) are used as markers of the nuclear periphery.Volz J, Carvalho TG, Ralph SA, Gilson P, Thompson J, Tonkin CJ, Langer C, Crabb BS, Cowman AF. Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum. Int J Parasitol. 2010 40:109-21. Copyright Elsevier 2010.
See original on MMP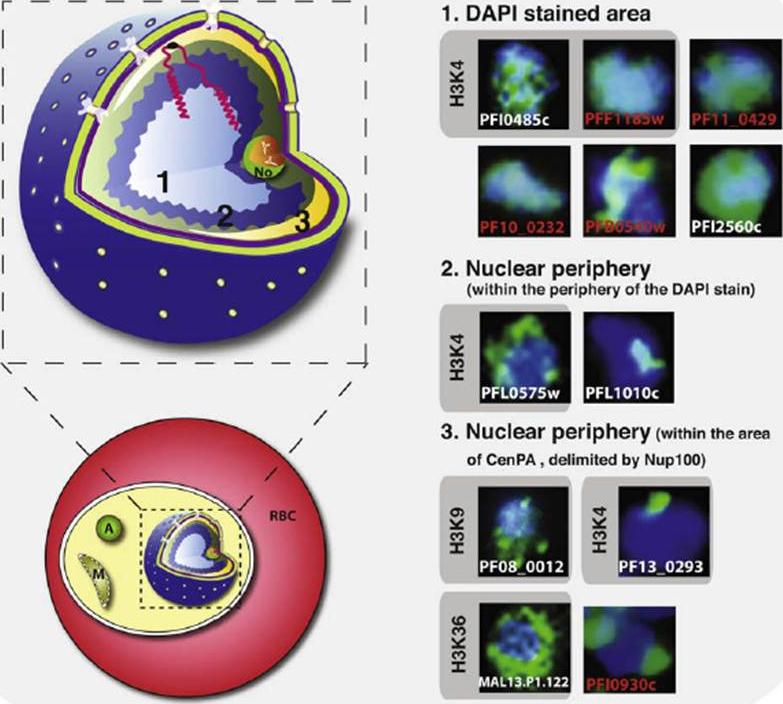
Schematic representation of the Plasmodium falciparum nuclear architecture. A schematic representation of an infected red blood cell (RBC) and an enlargement of the P. falciparum nucleus are shown. From the centre towards the periphery we have defined three distinct areas (1–3) in which nuclear proteins specifically distribute. A summary is shown in which trophozoite stages have been analyzed. Proteins, previously predicted to functionally interact, are shown in red. Proteins previously predicted for their histone target sites are indicated by grey boxes. A, apicoplast. M, mitochondria, No, nucleolus.Volz J, Carvalho TG, Ralph SA, Gilson P, Thompson J, Tonkin CJ, Langer C, Crabb BS, Cowman AF. Potential epigenetic regulatory proteins localize to distinct nuclear sub-compartments in Plasmodium falciparum. Int J Parasitol. 2010 40(1):109-21. Copyright Elsevier.
See original on MMPMore information
| PlasmoDB | PCHAS_0935100 |
| GeneDB | PCHAS_0935100 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PC000278.00.0, PC000615.01.0, PCAS_093510, PCHAS_093510 |
| Orthologs | PBANKA_0702900 , PF3D7_0827800 , PKNH_1321900 , PVP01_0506600 , PVX_088965 , PY17X_0703200 |
| Google Scholar | Search for all mentions of this gene |