PCHAS_0417200 nucleoporin NUP100/NSP100, putative (NUP100)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
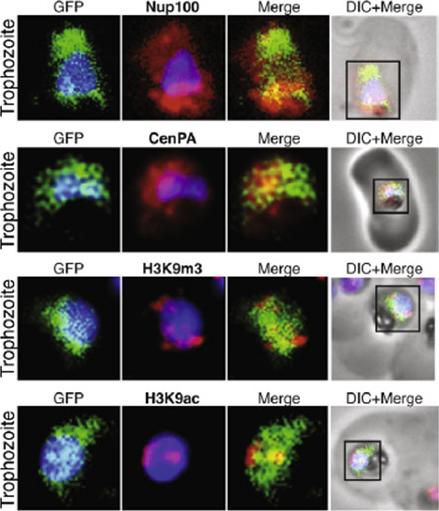
The protein encoded by the gene MAL13.P1.122 localize to the far nuclear periphery. MAL13P1.122 is a SET-domain protein (Histone-lysine N-methyltransferase), which localizes to the nuclear periphery surrounding the DAPI stained portion within the same region of CenPA (histone H3) and the histone mark H3K9m3. Co-localization studies by immunofluorescence. Nup100 (PFI0250c) and CenPA (histone H3 PF13_0185) are used as markers of the nuclear periphery.Volz J, Carvalho TG, Ralph SA, Gilson P, Thompson J, Tonkin CJ, Langer C, Crabb BS, Cowman AF. Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum. Int J Parasitol. 2010 40:109-21. Copyright Elsevier 2010.
See original on MMP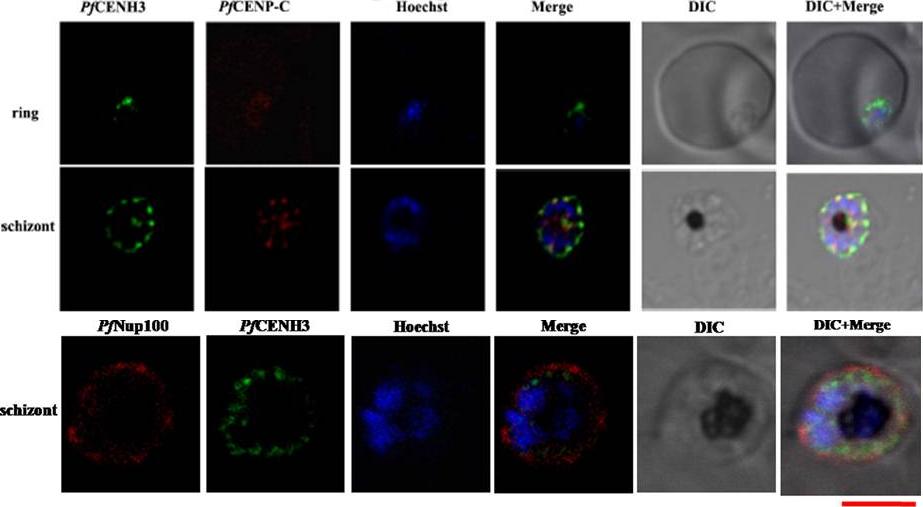
Upper: Colocalization of PfCENH3 with PfCENP-C. Confocal images showing the co-localization of the putative PfCENP-C (red) and PfCENH3 (green) at the nuclear periphery. The nucleus is stained with Hoechst (blue). Scale bar = 5 mm. Lower: The fluorescence pattern obtained on merging PfCENH3 and PfNup100with the nucleus depicts that PfCENH3 localizes to the nuclear periphery just outside the Hoechst stained nucleus but well within the PfNup100 that marks the outer nuclear boundaries.Verma G, Surolia N. Plasmodium falciparum CENH3 is able to functionally complement Cse4p and its, C-terminus is essential for centromere function. Mol Biochem Parasitol. 2013 Dec 6. [Epub ahead of print]
See original on MMP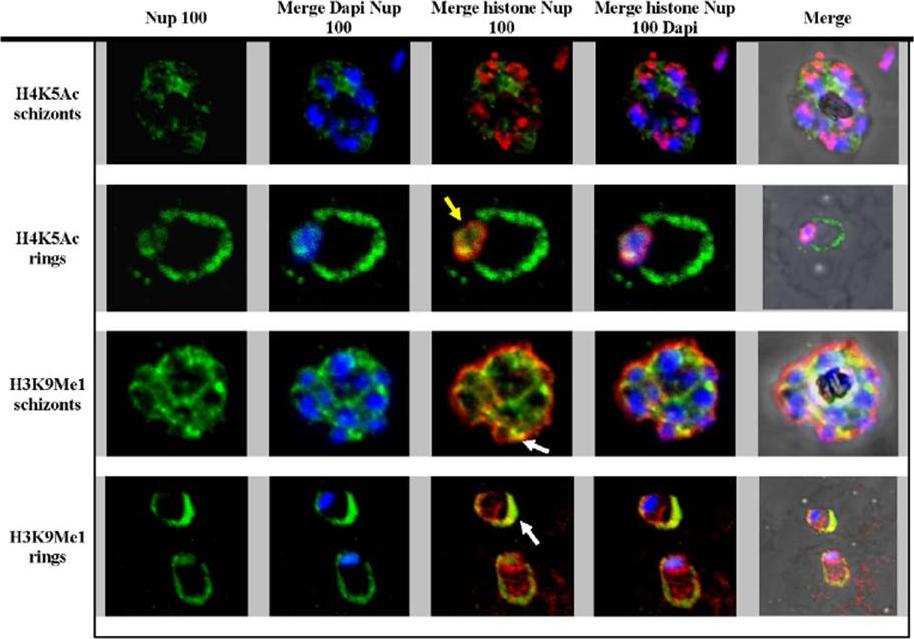
Immunofluorescence analysis of Nup 100. H3K9Me1, H4K5Ac and Nup 100 were stained red and green respectively. DAPI stained DNA blue. Yellow arrow indicates the co-localization of the H3K5Ac and Nup-100 in the ring stage parasites. White arrows indicate the co-localization of H3K9Me1 and Nup 100 outside the DAPI stained area, in the ring and schizont stage parasites.Luah YH, Chaal BK, Ong EZ, Bozdech Z. A moonlighting function of Plasmodium falciparum histone 3, mono-methylated at lysine 9? PLoS One. 2010 Apr 19;5(4):e10252
See original on MMP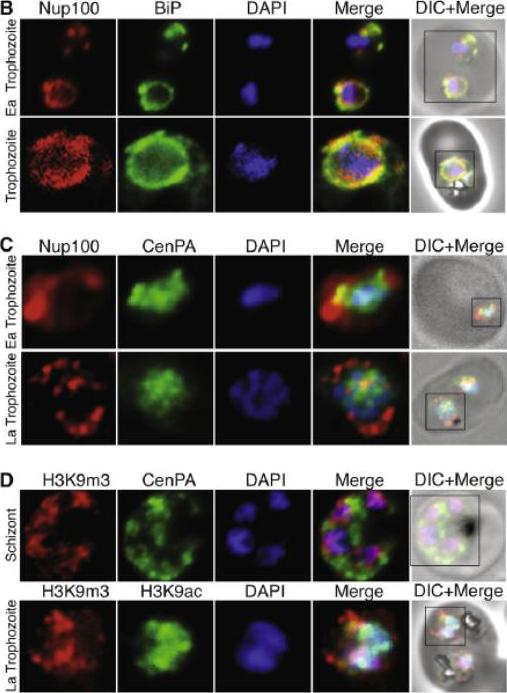
(B) Nucleoporin 100 (Nup100), a protein of the nuclear pore and BiP immunofluorescence analysis reveals the localization of Nup100 outside the DAPI stained area. Nup100 localizes in close proximity to BiP, a marker for the endoplasmic reticulum (ER) and does not overlap with DAPI, confirming that it defines the outer limits of the nucleus. (C) Localization of Nup100 and CenPA (histone 3) outside the DAPI stained area. CenPA and Nup100 do not co-localize; however, CenPA extends beyond the DAPI staining area of the nucleus suggesting that it is located within the periphery but inside the Nup100 delimited area of the nucleus (D) Localization of the histone marks H3K9m3 with CenPA and H3K9ac in the nucleus. The histone modification H3K9m3 silences transcription whereas H3K9ac is found associated with active transcription in general. H3K9m3 localized mostly to the nuclear periphery outside the DAPI stained area, which was consistent with the location of chromosome clusters containing silenced var genes. H3K9ac localized more with the DAPI stained area of the nucleus suggesting the DAPI stained area is broadly the transcriptionally active zone.Ea Trophozoite, Early Trophozoite; La trophozoite, Late Trophozoite.Volz J, Carvalho TG, Ralph SA, Gilson P, Thompson J, Tonkin CJ, Langer C, Crabb BS, Cowman AF. Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum. Int J Parasitol. 2010 40:109-21. Copyright Elsevier 2010.
See original on MMP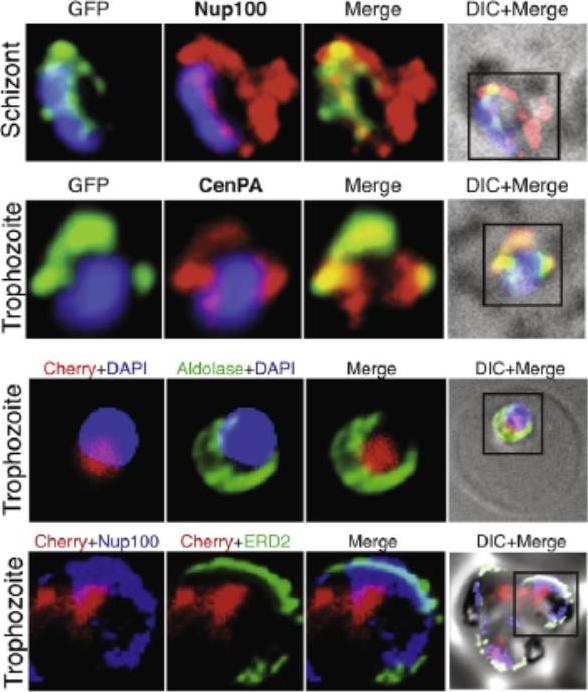
Nucleosome assembly protein PFI0930c localizes to the far nuclear periphery. (A,B) PFI0930c is either tagged with GFP or Cherry, respectively. Co-localization studies by immunofluorescence reveal its localization to the nuclear periphery, which is marked by CenPA (histone H3)and delimited by the nuclear envelope marker Nup100 (A), but not to the cytoplasmic portion of the cell marked by aldolase (PF14_0425) and ERD2 (PF13_0280) (B). Co-localisation of PFI0930c-cherry and aldolase, used as a cytoplasmic marker showed clear separation of nuclear and cytoplasmic regions in the P. falciparum cell. Co-localisation analysis with Nup100 and ERD2, a marker for the cis- and trans-Golgi showed the same overlapping localisation of PFI0930c-cherry and Nup100, both in an internal location relative to ERD2.Volz J, Carvalho TG, Ralph SA, Gilson P, Thompson J, Tonkin CJ, Langer C, Crabb BS, Cowman AF. Potential epigenetic regulatory proteins localise to distinct nuclear sub-compartments in Plasmodium falciparum. Int J Parasitol. 2010 40:109-21. Copyright Elsevier 2010.
See original on MMPMore information
| PlasmoDB | PCHAS_0417200 |
| GeneDB | PCHAS_0417200 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PC300215.00.0, PC300400.00.0, PC301578.00.0, PC301651.00.0, PC403721.00.0, PCAS_041720, PCHAS_041720 |
| Orthologs | PBANKA_0416300 , PF3D7_0905100 , PKNH_0702800 , PVP01_0703500 , PVX_098700 , PY17X_0419100 |
| Google Scholar | Search for all mentions of this gene |