PBANKA_0102900 transcription factor with AP2 domain(s), putative (SIP2)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Refractory |
20195509 | Theo Sanderson, Wellcome Trust Sanger Institute | |
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
| P. yoelii yoelii 17X |
Refractory |
RMgm-4381 | Imported from RMgmDB | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
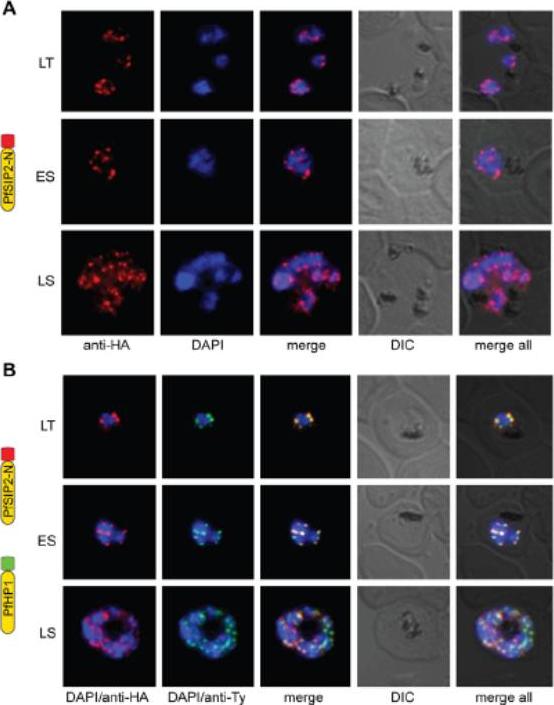
PfSIP2-N localizes to P. falciparum chromosome end clusters. (A) IFA detects discrete perinuclear PfSIP2-N-HA signals in late trophozoites (LT) and early (ES) and late (LS) schizont stage 3D7/SIP2-N-HA parasites. The expression cassette is schematically depicted on the left. (B) Co-localisation of PfSIP2-N with PfHP1-containing subtelomeric heterochromatin in late trophozoites (LT) and early (ES) and late (LS) schizonts in the double transgenic parasite line 3D7/SIP2-N-HA/HP1-Ty (over-expressing both proteins as epitope-tagged versions simultaneously). Expression cassettes are schematically depicted on the left. Indirect immunofluorescence (IFA) microscopy identified discrete PfSIP2-N-HA foci at the nuclear periphery with increasing numbers of foci in replicating stages (A) consistent with chromosome end clusters. PfHP1 is a heterochromatic marker. Both proteins co-localized at the nuclear periphery (B). Flueck C, Bartfai R, Niederwieser I, Witmer K, Alako BT, Moes S, Bozdech Z, Jenoe P, Stunnenberg HG, Voss TS. A major role for the Plasmodium falciparum ApiAP2 protein PfSIP2 in chromosome end biology. PLoS Pathog. 2010 6:e1000784.
See original on MMPMore information
| PlasmoDB | PBANKA_0102900 |
| GeneDB | PBANKA_0102900 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PB300561.00.0, PB405120.00.0, PBANKA_010290 |
| Orthologs | PCHAS_0103600 , PF3D7_0604100 , PKNH_1146400 , PVP01_1144800 , PVX_113370 , PY17X_0104500 |
| Google Scholar | Search for all mentions of this gene |