PVX_082675 merozoite surface protein 7 (MSP7)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. falciparum 3D7 |
Possible |
20472690 | Theo Sanderson, Wellcome Trust Sanger Institute | |
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. falciparum 3D7 | Asexual |
No difference |
20472690 | Theo Sanderson, Wellcome Trust Sanger Institute |
Imaging data (from Malaria Metabolic Pathways)
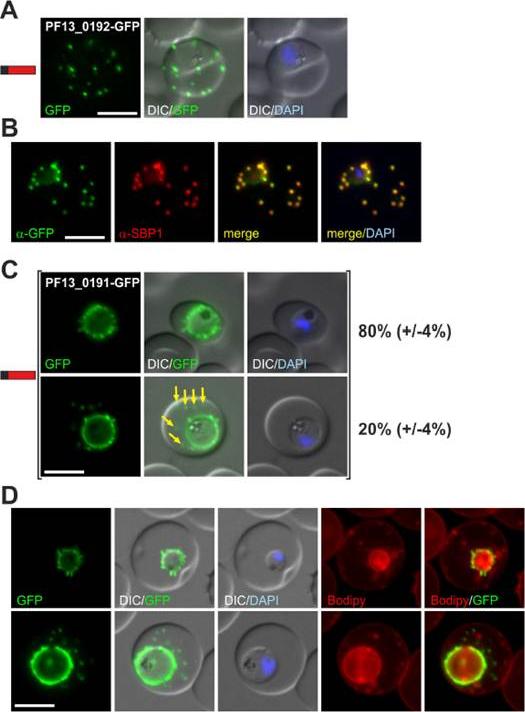
(A) Fluorescence pattern of PF13_0192-GFP. (B) Co-localisation IFA of PF13_0192-GFP with SBP1. (C) Fluorescence pattern of PF13_0191-GFP. Two panels are shown to demonstrate cells with (yellow arrows) and without additional foci of fluorescence in the host cell (ratio indicated in %, at least 50 cells were analysed on 3 occasions, standard deviation in brackets). (D) Bodipy-TR-C5-ceramide (Bodipy) stained parasites expressing PF13_0191-GFP. Protein structure in A and C indicated as in Figure 1. Nuclei were stained with DAPI. Size bars: 5 mm. PF13_0192-GFP was exported to foci in the host cell (A) that were confirmed to be Maurer’s clefts (B). PF13_0191-GFP localized to foci and mobile protrusions at the parasite periphery and in 20% (+/24%) of cells was also found in usually multiple mobile foci in the host cell with no apparent contact to the parasite periphery.Heiber A, Kruse F, Pick C, Grüring C, Flemming S, Oberli A, Schoeler H, Retzlaff S, Mesén-Ramírez P, Hiss JA, Kadekoppala M, Hecht L, Holder AA, Gilberger TW, Spielmann T. Identification of New PNEPs Indicates a Substantial Non-PEXEL Exportome and Underpins Common Features in Plasmodium falciparum Protein Export. PLoS Pathog. 2013 9(8):e1003546.
See original on MMPMore information
| PlasmoDB | PVX_082675 |
| GeneDB | PVX_082675 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | Pv082675 |
| Orthologs | PF3D7_1334300 , PKNH_1266100 , PVP01_1219800 |
| Google Scholar | Search for all mentions of this gene |