PVP01_0504300 conserved Plasmodium protein, unknown function
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
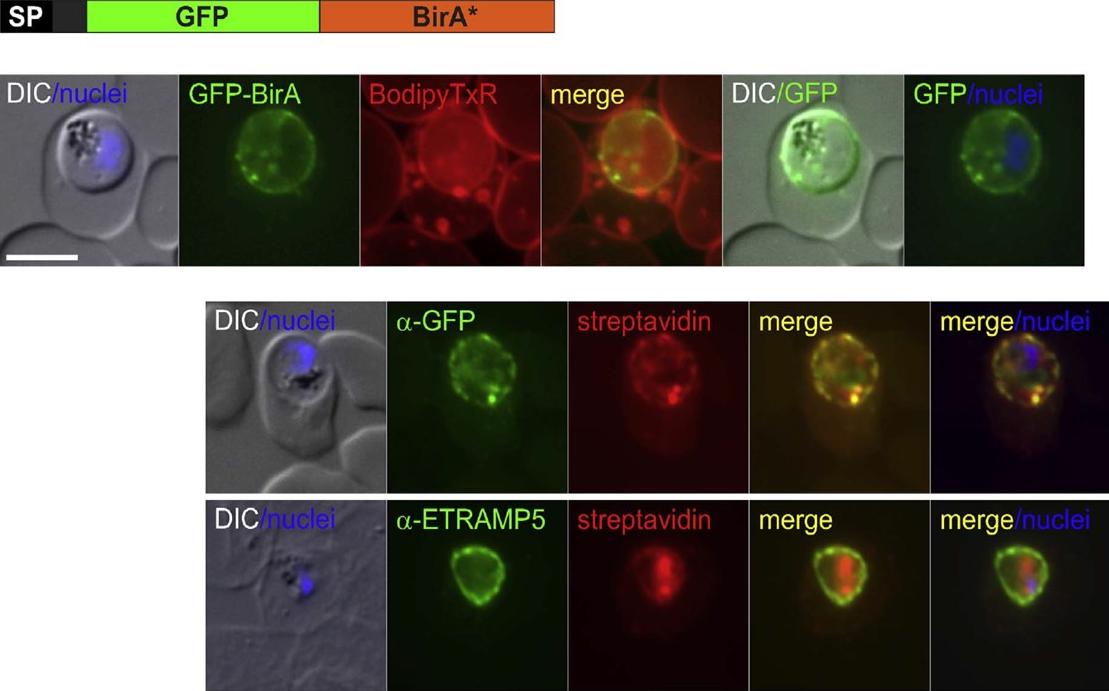
The SP-GFP-BirA* construct is located in the PV and is able to biotinylate proteins in this compartment. Immune fluorescence assay (IFA) of SP-GFP-BirA* expressing parasites grown in the presence of biotin show co-localisation of biotinylated protein (detected using streptavidin) with GFPBirA* (upper panel) and ETRAMP5 (lower panel). To test whether BirA* can biotinylate proteins in the PV we grew the SP-GFP-BirA* expressing parasites in the presence of biotin. Immuno and streptavidin-fluorescence assays showed that this resulted in biotinylation of proteins in the parasite periphery and that the detected biotin co-localized with the SP-GFP-BirA* construct and with the PVM marker ETRAMP5.Khosh-Naucke M, Becker J, Mesén-Ramírez P, Kiani P, Birnbaum J, Fröhlke U, Jonscher E, Schlüter H, Spielmann T. Identification of novel parasitophorous vacuole proteins in P. falciparum parasites using BioID. Int J Med Microbiol. 2017 Jul 27. [Epub ahead of print]
See original on MMPMore information
| PlasmoDB | PVP01_0504300 |
| GeneDB | PVP01_0504300 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | null |
| Orthologs | PF3D7_0830400 , PKNH_1324200 , PVX_088855 |
| Google Scholar | Search for all mentions of this gene |