PF3D7_1407100 rRNA 2'-O-methyltransferase fibrillarin, putative (NOP1)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
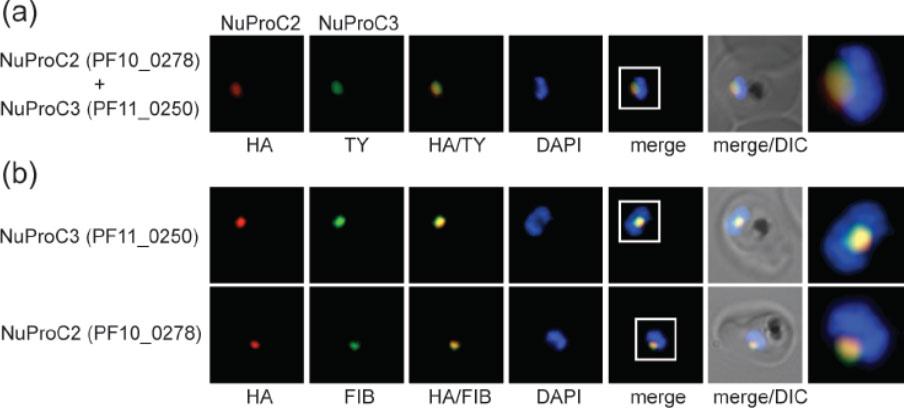
Identification of two novel P. falciparum nucleolar proteins. (a) IFA colocalisation of NuProC2-3xHA and NuProC3-2xTy in a double transgenic line. Epitope-tagged proteins were detected using anti-HA (red) and anti-Ty (green) antibodies. (b) NuProC2-3xHA and NuProC3-3xHA co-localize with nucleolar fibrillarin. HA-tagged proteins were detected in single transgenic lines using anti-HA antibodies (red). Fibrillarin (PfFIB) was visualized using an anti-human fibrillarin antibody.Oehring SC, Woodcroft BJ, Moes S, Wetzel J, Dietz O, Pulfer A, Dekiwadia C, Maeser P, Flueck C, Witmer K, Brancucci NM, Niederwieser I, Jenoe P, Ralph SA, Voss TS. Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol. 2012 Nov 26;13(11):R108.
See original on MMP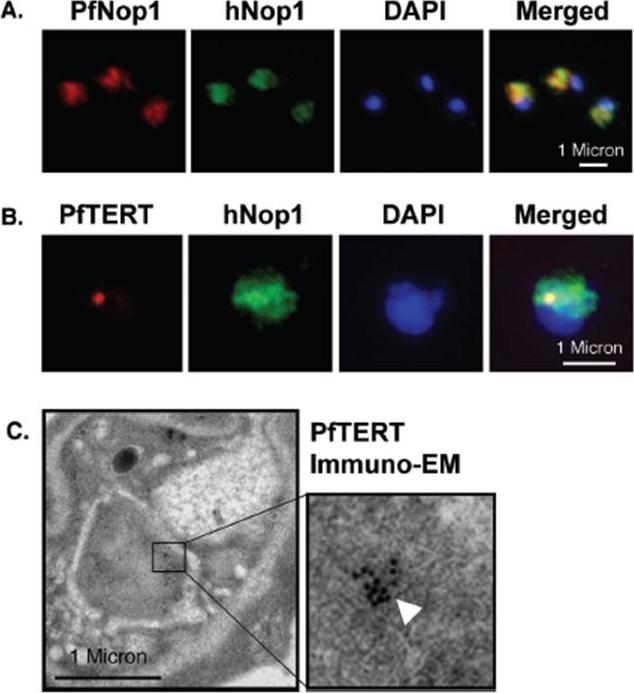
PfTERT localizes in a sub-compartment of the nucleolus. (A) IF with an anti P. falciparum Nop1 serum (red) shows the nucleolus as a hat-like structure in the nucleus of a trophozoite stage. An anti-human Nop1 serum (green) recognizes exactly the same sub-nuclear region. (B) Double-labelling reveals that PfTERT (red) co-localizes with part of the nucleolus (green); yellow spot in the merge image. (C) Immuno-EM shows that PfTERT (white arrow head) localizes to a peripheral region of the nucleus. In (A and B), DNA was stained with DAPI (blue).Figueiredo LM, Rocha EP, Mancio-Silva L, Prevost C, Hernandez-Verdun D, Scherf A. The unusually large Plasmodium telomerase reverse-transcriptase localizes in a discrete compartment associated with the nucleolus. Nucleic Acids Res. 2005 33:1111-22. by permission of Oxford University Press.
See original on MMP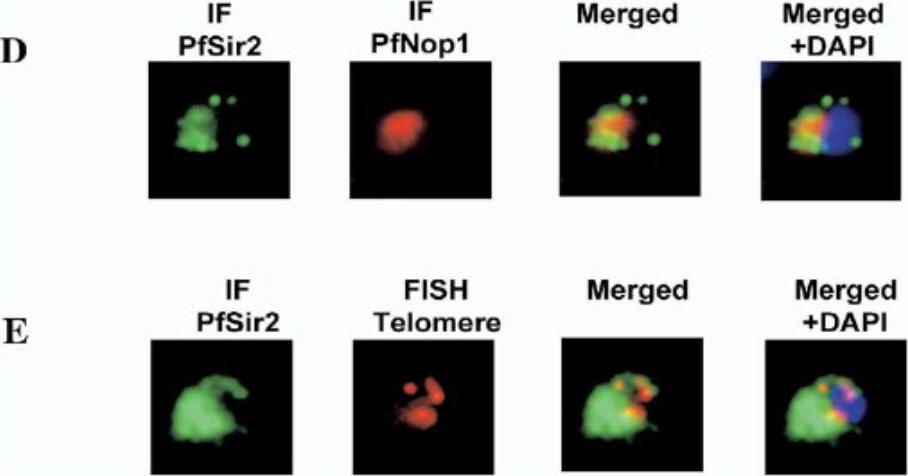
(D) IF using the PfNop1 and PfSir2 antibodies. The PfSir2 antibody is visualized in red and the PfNop1 antibody in green using Alexa 488. (E) Simultaneous IF and FISH analysis. Immunolocalization (IF) of PfSir2 was performed on young trophozoite parasites using an Alexa 488-conjugated secondary antibody (green). FISH analysis was performed simultaneously using a telomeric oligonucleotide probe conjugated with Cy3 (red). The nuclei were stained with DAPI. Protein localizes to telomeric foci. anti-PfSir2 antibody signal is highly concentrated at one pole of the nucleus.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mâncio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005 121:25-36. Copyright Elsevier 2009.
See original on MMP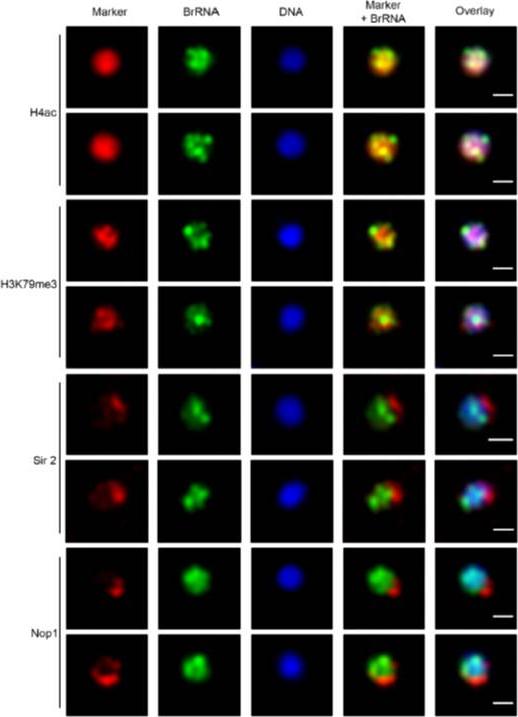
Transcription sites form a distinct compartment in the nucleus of P. falciparum. Immunofluorescence assay for nascent RNAs of ring stage parasites labeled with BrUTP (BrRNA, green) and the compartments of acetylated H4 (H4ac, active chromatin); a proposed histone marker of nascent transcription (H3K79me3; PfSir2A (silencing compartment); and Nop1 (nucleolar compartment). DNA was labeled by DAPI, in blue. Bars, 1 mm.Moraes CB, Dorval T, Contreras-Dominguez M, Dossin Fde M, Hansen MA, Genovesio A, Freitas-Junior LH. Transcription sites are developmentally regulated during the asexual cycle of Plasmodium falciparum. PLoS One. 2013;8(2):e55539.
See original on MMP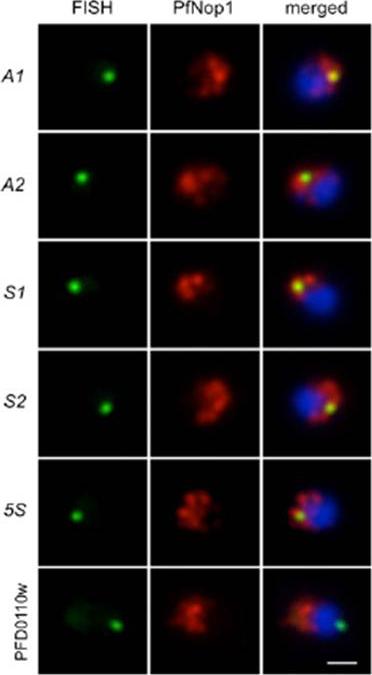
Immunofluorescence analysis using antibodies against the nucleolar protein PfNop1 (red) combined with DNA FISH (green) for individual rRNA genes (A1, A2, S1, S2, and 5S) and a subtelomeric unrelated gene (PFD0110w) on chromosome 4. The percentage of signals that localized in or near the nucleolus and the number of nuclei analyzed are as follows: A1 93.2%, n = 73; A2 90.5%, n = 95; S1 92.3%, n = 91; S2 90.0%, n = 80; 5S 90.6%, n = 128; PFD0110w 24.6%, n = 57. Parasites are in ring stage, and nuclear DNA is stained with DAPI (blue). (Scale bar, 1 μm.Mancio-Silva L, Zhang Q, Scheidig-Benatar C, Scherf A. Clustering of dispersed ribosomal DNA and its role in gene regulation and chromosome-end associations in malaria parasites. Proc Natl Acad Sci U S A. 2010 107(34):15117-22.
See original on MMP
Upper 2 rows: Immunofluorescence analysis of PfPol I (red). Ring-stage parasites show a half-moon perinuclear nucleolus. In trophozoites, PfPol I shows an additional diffuse pattern in the nucleoplasm and cytoplasm. In schizont segmented forms, PfPol I appears to relocalize at the nuclear periphery. Parasite nuclei are shown by DAPI staining (blue). (Scale bars, 2 μm.)Lower 3 rows: Double-labeling immunofluorescence assays using rabbit anti-PfPol I and rat anti-PfNop1 (A) or anti-PfNop5 (B) antibodies, showing localization of the nucleolus at periphery of nucleus, forming a “hat-like” structure. (C) RNA FISH (using a 18S universal probe that hybridizes to all rRNAs produced in the parasite, green) combined with immunofluorescence microscopy (using the anti-PfPolI antibody, red), demonstrating that the region detected by the antibodies corresponds to the nucleolus of the parasite. In all images, parasites are in the ring stage, and the nuclear DNA is stained with DAPI (blue). (Scale bars, 1 μm.)Mancio-Silva L, Zhang Q, Scheidig-Benatar C, Scherf A. Clustering of dispersed ribosomal DNA and its role in gene regulation and chromosome-end associations in malaria parasites. Proc Natl Acad Sci U S A. 2010 107:15117-22.
See original on MMP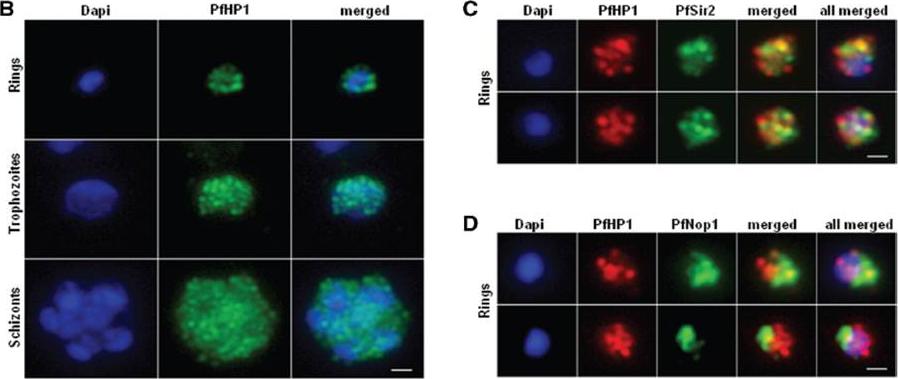
Nuclear expression and cellular localization of PfHP1. (B) IFA analysis of PfHP1 (green) throughout the parasite asexual blood stage cycle (ring, trophozoite and schizont stages). (C) In ring stage, partial co-localization between PfHP1 (red) and PfSir2 (green) antibodies (yellow spots) is observed. (D) Co-localization with a nucleolar marker PfNop1 shows minimal overlap of PfHP1 with this compartment. Scale bars represent 1 mm.Pérez-Toledo K, Rojas-Meza AP, Mancio-Silva L, Hernández-Cuevas NA, Delgadillo DM, Vargas M, Martínez-Calvillo S, Scherf A, Hernandez-Rivas R. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009 37(8):2596-606
See original on MMP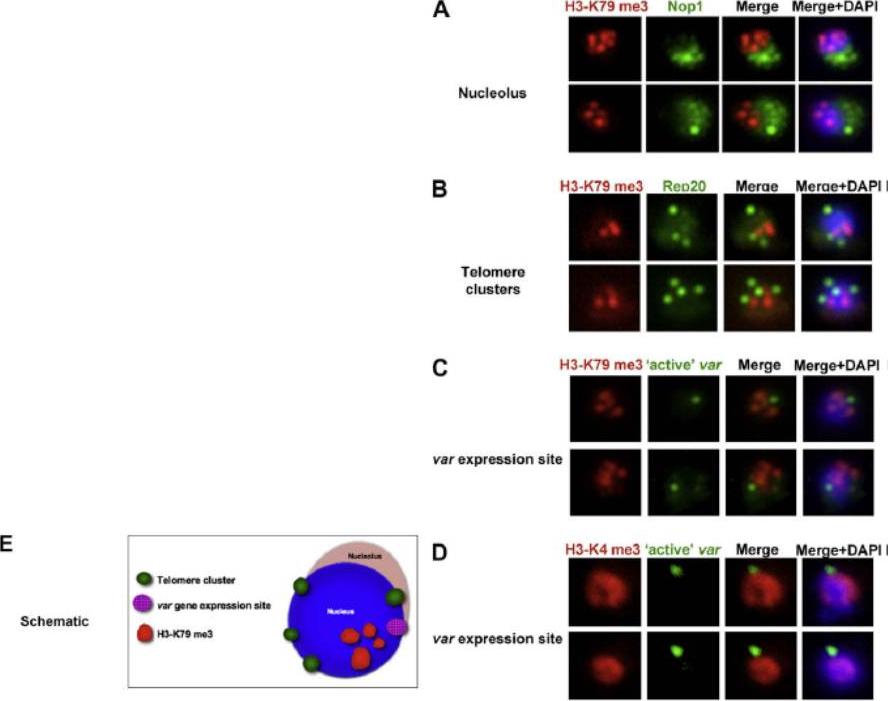
Co-localisation of H3-K79me3 with known cellular compartments. A. Nucleolus: Double labelling IFAs with antibodies against H3-K79me3 and nucleolus marker, PfNop1. B. Telomere clusters: IFeFISH analysis between H3-K79me3 (red) and Rep20 (green). C. Active var gene: IF-FISH analysis between H3-K79me3 (red) and active var gene (green). D. Active var gene colocalisation with activating mark, H3-K4me3. IF-FISH analysis between H3-K4me3 (red) and active var gene (green). DAPI stained nucleus in blue. Font colour represents corresponding fluorescein labelled antibody. Scale bar depicts 1 mm. E. Schematic of the dynamic compartmen-talisation of the P. falciparum nucleus. The distinct sub-nuclear distribution of histone methylation modification, taking example of H3-K79me3 as observed in this study. Known compartments compatible with active transcription are indicated. H3-K79me3 occupies a distinct zone.Issar N, Ralph SA, Mancio-Silva L, Keeling C, Scherf A. Differential sub-nuclear localisation of repressive and activating histone methyl modifications in P. falciparum. Microbes Infect. 2009 11:403-7.
See original on MMP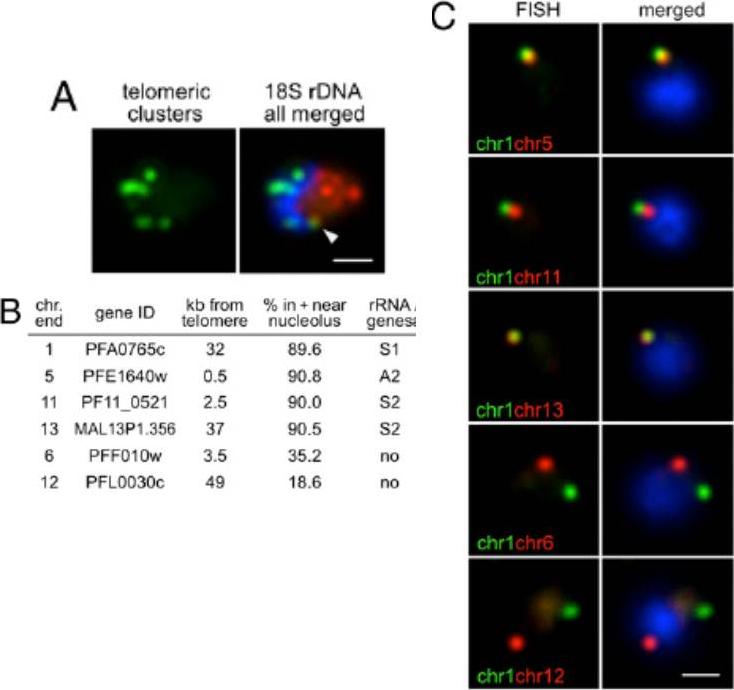
Nucleolar clustering of rDNA determines preferential association of rDNA-carrying chromosome ends. (A) Two-color DNA FISH of telomeric clusters (rep20, green) and 18S rDNA (red). The 18S rDNA FISH probe simultaneously detects the five rDNA units, which are clustered at one pole of the nucleus opposing the telomere clusters. White arrow points to the potential nucleolus-associated telomeric cluster. (B) Table presenting relative nuclear position of chromosome-ends (all genes encode PfEMP1) used in C determined by immuno-FISH using PfNop1 antibodies. (C) Two-color DNA FISH analysis of chromosome end pairs. Chromosome end 1 is labeled in green, and chromosome ends 5, 11, 13, 6, and 12 are shown in red. In all images, the parasites are in ring-stage, and nuclei are shown by DAPI staining (blue). (Scale bar, 1 μm).Mancio-Silva L, Zhang Q, Scheidig-Benatar C, Scherf A. Clustering of dispersed ribosomal DNA and its role in gene regulation and chromosome-end associations in malaria parasites. Proc Natl Acad Sci U S A. 2010 107:15117-22.
See original on MMPMore information
| PlasmoDB | PF3D7_1407100 |
| GeneDB | PF3D7_1407100 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PF14_0068 |
| Orthologs | PBANKA_1035100 , PCHAS_1035900 , PKNH_1351000 , PVP01_1341600 , PVX_086075 |
| Google Scholar | Search for all mentions of this gene |