PF3D7_1346300 DNA/RNA-binding protein Alba 2 (ALBA2)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
| P. berghei ANKA |
Possible |
PlasmoGEM (Barseq) | PlasmoGEM | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
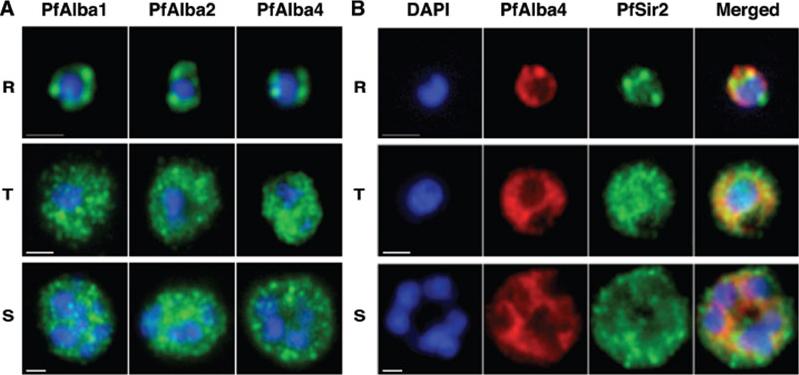
PfAlbas localize to perinuclear foci in ring stages and expand to the cytoplasm in mature forms. (A) Immunofluorescence analysis of PfAlba1, PfAlba2 and Pfalba4 distribution throughout the parasite life cycle (Rings, Trophozoites, Schizonts). The anti-PfAlba antibodies (Ab 74–2 for PfAlba1; Ab 233 for PfAlba2; Ab 237–2 for PfAlba4) were used at 5.5, 4.5 and 8 mg/ml, respectively. Anti-rabbit Alexa-488 secondary antibodies (Molecular Probes) were used at a dilution of 1:500. Scale bars represent 1 mm. (B) Immunofluorescence analysis of 3D7 Rings, Trophozoites and Schizonts stained with anti-PfAlba4 (red) and anti-PfSir2 (green) antibodies. Scale bar represents 1 mm.Chêne A, Vembar SS, Rivière L, Lopez-Rubio JJ, Claes A, Siegel TN, Sakamoto H, Scheidig-Benatar C, Hernandez-Rivas R, Scherf A. PfAlbas constitute a new eukaryotic DNA/RNA-binding protein family in malaria parasites. Nucleic Acids Res. 2012 40(7):3066-77
See original on MMPMore information
| PlasmoDB | PF3D7_1346300 |
| GeneDB | PF3D7_1346300 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | MAL13P1.233 |
| Orthologs | PBANKA_1359200 , PCHAS_1363800 , PKNH_1254800 , PVP01_1208500 , PVX_083215 , PY17X_1364900 |
| Google Scholar | Search for all mentions of this gene |