PF3D7_1332000 syntaxin, Qa-SNARE family (SYN5)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
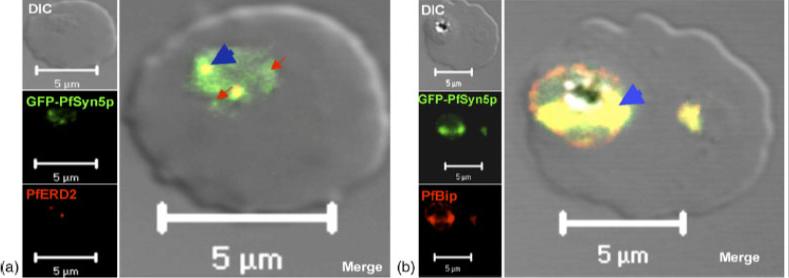
Subcellular localization of GFP-PfSyn5p. The GFP-expressing parasites were fixed and stained with anti-PfERD2 or anti-PfBip antibodies. As indicated by blue arrowheads, the fusion protein GFP-PfSyn5p (green) co-localizes with the cis-Golgi marker PfERD2 (red, panel a) and with the ER marker PfBip (red, panel b). The inserted red arrows show GFP fluorescence in locations different from cis-Golgi or ER compartments.Ayong L, Pagnotti G, Tobon AB, Chakrabarti D. Identification of Plasmodium falciparum family of SNAREs. Mol Biochem Parasitol. 2007 152:113-22. Copyright Elsevier 2009.
See original on MMPMore information
| PlasmoDB | PF3D7_1332000 |
| GeneDB | PF3D7_1332000 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | MAL13P1.169 |
| Orthologs | PBANKA_1346800 , PCHAS_1351400 , PKNH_1268600 , PVP01_1222700 , PVX_082530 , PY17X_1351800 |
| Google Scholar | Search for all mentions of this gene |