PF3D7_1214100 GPI ethanolamine phosphate transferase 3, putative (PIGO)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
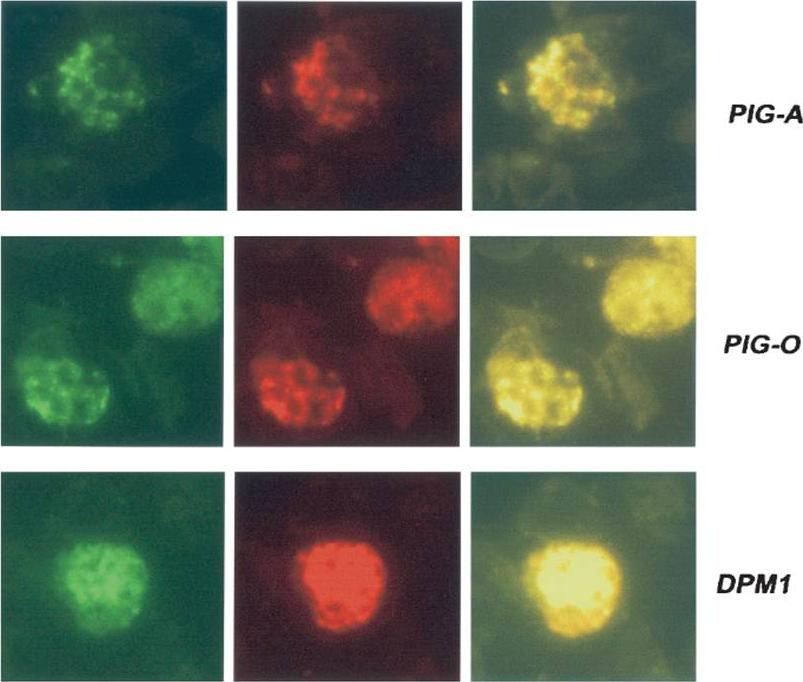
Detection of proteins in blood stage parasites was performed using indirect fluorescent-antibody tests. Thin films of parasites at mature stages were fixed in cold acetone and incubated sequentially with 1/80 dilutions of sera from peptide-immunized mice plus rabbit anti-ERC1 (1:200) to determine localization to the parasite ER. These slides were washed and incubated with an appropriate mixture of fluorescein-conjugated anti-mouse and rhodamine-conjugated anti-rabbit (1:1.000) antibodies. Slides were photographed under appropriate illumination for fluorescein isothiocyanate (first column, showing PIG-A, PIG-O, or DPM1 localization) and rhodamine (second column, showing ERC1 localization), and photographs were overlaid (third column).Delorenzi M, Sexton A, Shams-Eldin H, Schwarz RT, Speed T, Schofield L. Genes for glycosylphosphatidylinositol toxin biosynthesis in Plasmodium falciparum. Infect Immun. 2002 70:4510-22. PMID:
See original on MMPMore information
| PlasmoDB | PF3D7_1214100 |
| GeneDB | PF3D7_1214100 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | 2277.t00138, MAL12P1.137, PFL0685w |
| Orthologs | PBANKA_1429900 , PCHAS_1431800 , PKNH_1433600 , PVP01_1432900 , PVX_123395 , PY17X_1432100 |
| Google Scholar | Search for all mentions of this gene |