PF3D7_1200400 erythrocyte membrane protein 1, PfEMP1 (VAR)
Disruptability [+]
| Species | Disruptability | Reference | Submitter |
|---|---|---|---|
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
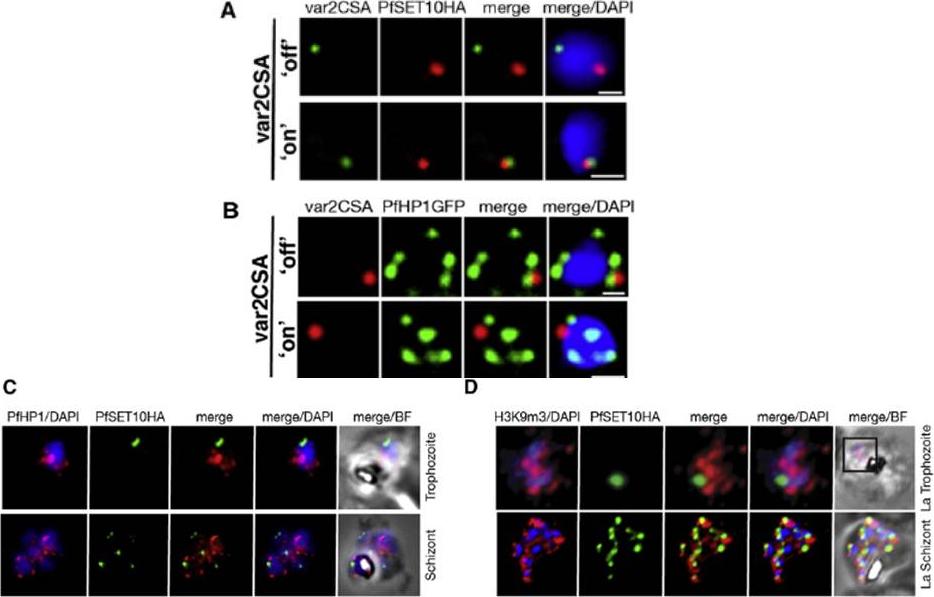
A. IFA-FISH of 3D7SET10-HA, in which var2csa (PFL0030c) activation has been selected by CSA adherence (‘‘on’’) (bottom panel). Unselected 3D7SET10-HA (top panel) has var2csa ‘‘off.’’ The localization of var2csa (green) and PfSET10 (red) is shown with both merged with the nucleus stained by DAPI (blue). Scale bars are equivalent to 0.5 mm. B. PfHP1 does not localize with active var2csa (‘‘on’’). The var2csa gene (red) location is shown with respect to PfHP1 protein (green). C. PfHP1 (red) and D. histone mark H3K9me3 (red,), known markers of silent var gene clusters and PfSET10 (green), do not colocalize in the nucleus stained with DAPI (blue).Volz JC, Bártfai R, Petter M, Langer C, Josling GA, Tsuboi T, Schwach F, Baum J, Rayner JC, Stunnenberg HG, Duffy MF, Cowman AF. PfSET10, a Plasmodium falciparum Methyltransferase, Maintains the Active var Gene in a Poised State during Parasite Division. Cell Host Microbe. 2012 11(1):7-18.
See original on MMPMore information
| PlasmoDB | PF3D7_1200400 |
| GeneDB | PF3D7_1200400 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | 2277.t00004, MAL12P1.4, PFL0020w |
| Orthologs | |
| Google Scholar | Search for all mentions of this gene |