PF3D7_0631500 exported protein family 3 (EPF3)
Disruptability [+]
| Species | Disruptability | Reference | Submitter |
|---|---|---|---|
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
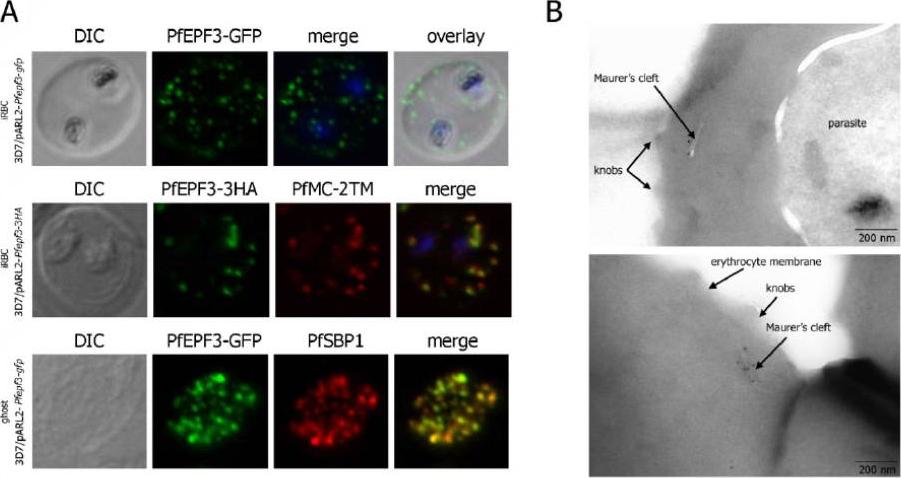
Localization and topology of the PfEPF3 proteins. A. Fluorescent patterns of iRBCs infected by 3D7/pARL2-Pfepf3-gfp (live imaging) and 3D7/pARL2-Pfepf3-3HA (immunodetection using anti-HA antibodies, in green, and anti-PfMC-2TM antibodies, in red) and of resealed ghosts from 3D7/pARL2-Pfepf3-gfp iRBCs (GFP fluorescence, in green, and immunodetection using anti-SBP1 antibodies, in red). RBCs and ghost preparations were incubated with DAPI for nucleus labelling. B. Immunoelectron microscopy using anti-GFP antibodies.Mbengue A, Audiger N, Vialla E, Dubremetz JF, Braun-Breton C. Novel Plasmodium falciparum Maurer's clefts protein families implicated in the release of infectious merozoites. Mol Microbiol. 2013 88(2):425-42 PMID: .
See original on MMPMore information
| PlasmoDB | PF3D7_0631500 |
| GeneDB | PF3D7_0631500 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | 2270.t00310, MAL6P1.14, PFF1530c |
| Orthologs | |
| Google Scholar | Search for all mentions of this gene |