PF3D7_0532100 early transcribed membrane protein 5 (ETRAMP5)
Disruptability [+]
| Species | Disruptability | Reference | Submitter |
|---|---|---|---|
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
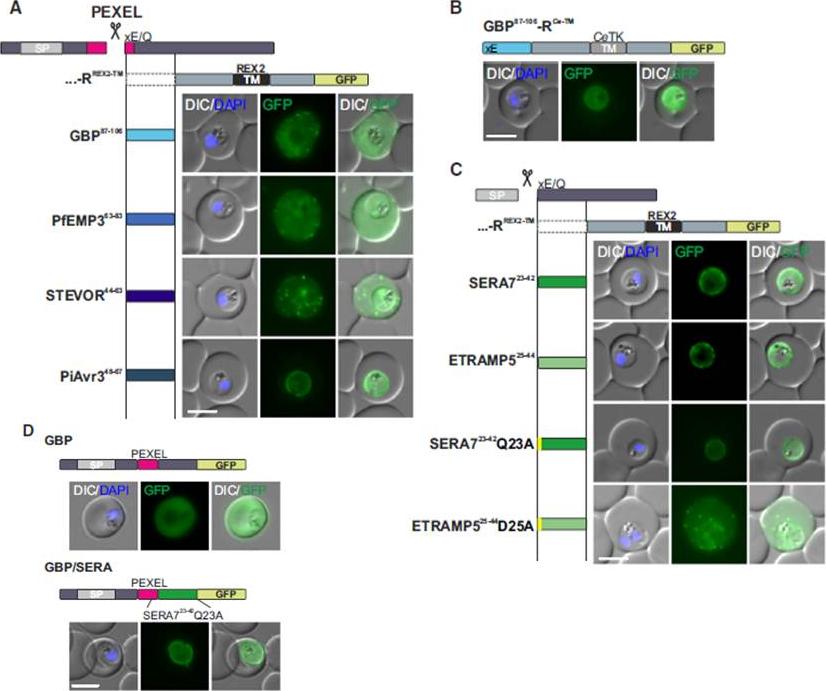
Mature PEXEL N Termini Promote Export of RREX2-TM (A–C) Images of live P. falciparum parasites expressing RREX2-TM fused with the mature N termini of PEXEL proteins (A), GBP87–106-RCe-TM (B), or RREX2-TM fused with the mature N termini of nonexported secretory proteins (C). The position of the appended region in the original protein is shown above each panel.(D) Images of live P. falciparum parasites expressing truncated GBP fused to GFP (GBP, top) or GBP-GFP with the mature N terminus of SERA7Q23A after the PEXEL (GBP/SERA, bottom). Although these N termini contain only the last of the conserved PEXEL residues (PEXEL position 5) and the nonconserved position 4, they promoted export of the reporter into the host cell (A). GFP fluorescence was detected in the erythrocyte cytosol and the Maurer’s clefts.Grüring C, Heiber A, Kruse F, Flemming S, Franci G, Colombo SF, Fasana E,Schoeler H, Borgese N, Stunnenberg HG, Przyborski JM, Gilberger TW, Spielmann T. Uncovering common principles in protein export of malaria parasites. Cell Host Microbe. 2012 12(5):717-29.
See original on MMP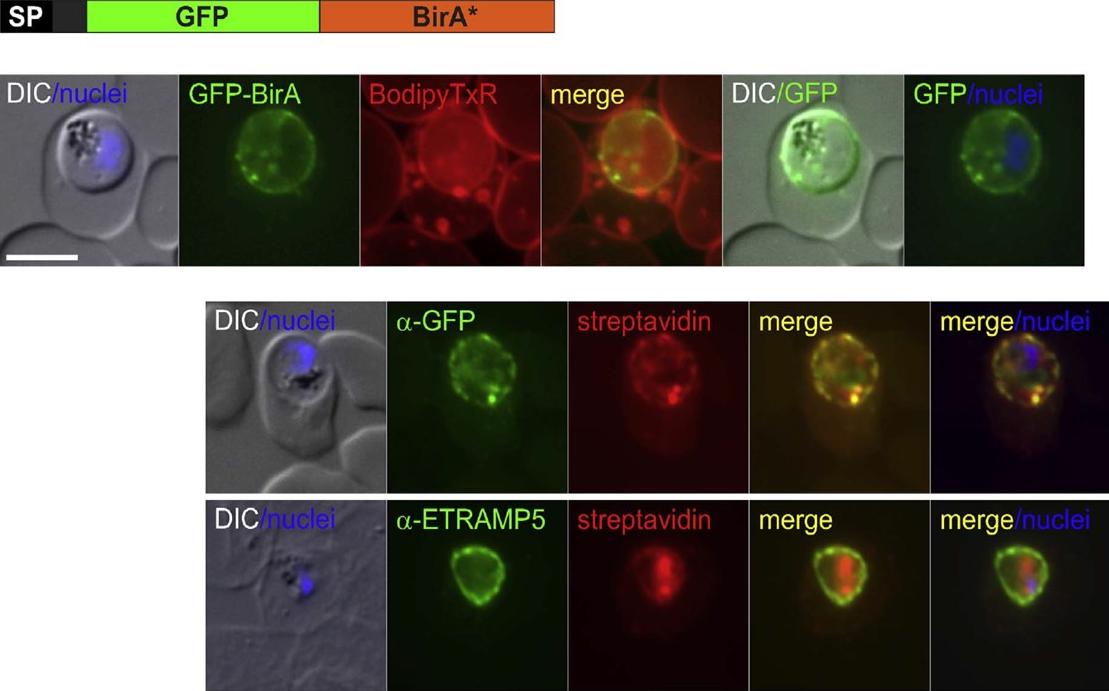
The SP-GFP-BirA* construct is located in the PV and is able to biotinylate proteins in this compartment. Immune fluorescence assay (IFA) of SP-GFP-BirA* expressing parasites grown in the presence of biotin show co-localisation of biotinylated protein (detected using streptavidin) with GFPBirA* (upper panel) and ETRAMP5 (lower panel). To test whether BirA* can biotinylate proteins in the PV we grew the SP-GFP-BirA* expressing parasites in the presence of biotin. Immuno and streptavidin-fluorescence assays showed that this resulted in biotinylation of proteins in the parasite periphery and that the detected biotin co-localized with the SP-GFP-BirA* construct and with the PVM marker ETRAMP5.Khosh-Naucke M, Becker J, Mesén-Ramírez P, Kiani P, Birnbaum J, Fröhlke U, Jonscher E, Schlüter H, Spielmann T. Identification of novel parasitophorous vacuole proteins in P. falciparum parasites using BioID. Int J Med Microbiol. 2017 Jul 27. [Epub ahead of print]
See original on MMPMore information
| PlasmoDB | PF3D7_0532100 |
| GeneDB | PF3D7_0532100 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | MAL5P1.312, PFE1590w |
| Orthologs | |
| Google Scholar | Search for all mentions of this gene |