PF3D7_0516100 cation-transporting ATPase 1 (ATPase1)
Disruptability [+]
| Species | Disruptability | Reference | Submitter |
|---|---|---|---|
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
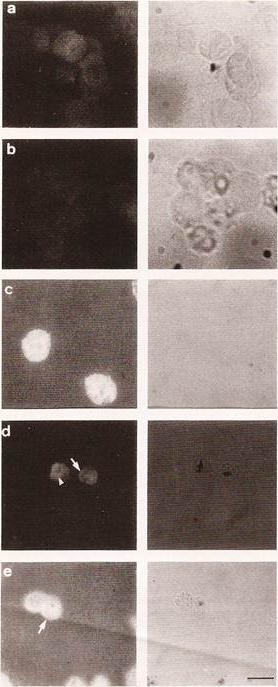
Localization of ATPase 1 by IFA. Abs to two regions of derived amino acid sequence from ATPase 1(anti-pep1 and anti-pep2) were applied to slides from isolate T9/96. Figures show dark filed (fluorescence) and bright filed views. Hemozoin pigment in bright field views corresponds to the location of parasitized red blood cells. a) Negative control (no FITC-conjugated Abs). b) negative control (no primary Abs). c) anti-Band 3 mAbs; d) anti pep1 Abs. e) anti-pep2 Abs. The arrows indicate fluorescence in the region of the parasite plasma membrane/parasitophorous vacuole, and the arrowhead indicates staining internal to the parasite. Bar: 10 mm.Krishna S, Cowan G, Meade JC, Wells RA, Stringer JR, Robson KJ. A family of cation ATPase-like molecules from Plasmodium falciparum. J Cell Biol. 1993 120:385-98.
See original on MMPMore information
| PlasmoDB | PF3D7_0516100 |
| GeneDB | PF3D7_0516100 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | MAL5P1.161, PFE0805w |
| Orthologs | PKNH_1029600 , PVP01_1030000 , PVX_097770 |
| Google Scholar | Search for all mentions of this gene |