PF3D7_0401800 Plasmodium exported protein (PHISTb), unknown function (PfD80)
Disruptability [+]
| Species | Disruptability | Reference | Submitter |
|---|---|---|---|
| P. falciparum 3D7 |
Refractory |
18614010 | Theo Sanderson, Wellcome Trust Sanger Institute |
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
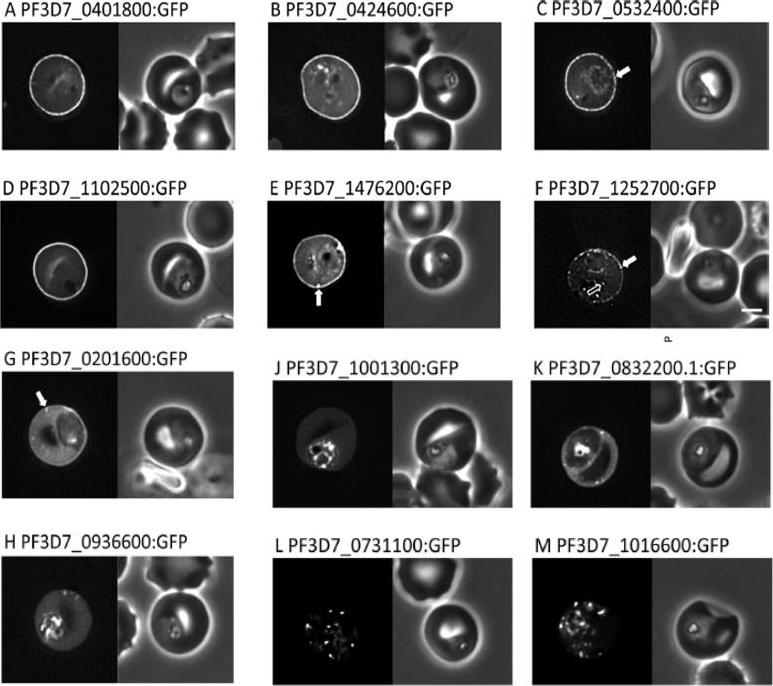
Localisation of PHIST:GFP proteins. The left- and right-hand images show GFP localisation and a phase contrast image, respectively. White filled arrows: peripheral GFP puncta; White unfilled arrow: GFP puncta in host erythocyte cytosol. Scale bar, 2 mm. GFP-tagged PF3D7_0401800, PF3D7_0424600, PF3D7_0532400, PF3D7_1102500 and PF3D7_1476200 were all exported to the host cell and displayed a striking localisation at the edge of the host erythrocyte (1A–E), PF3D7 1252700 was clearly peripheral in the host erythrocyte (F). PF3D7_0201600 (G) shows weak accumulation at the erythrocyte periphery, but the majority of the protein was localised in the RBC cytosol. PF3D7_0936600 was localised in the erythrocyte cytosol, not at the host cell periphery (H).Tarr SJ, Moon RW, Hardege I, Osborne AR. A conserved domain targets exported PHISTb family proteins to the periphery of Plasmodium infected erythrocytes. Mol Biochem Parasitol. 2014 196(1):29-40 PMID: 25106850
See original on MMP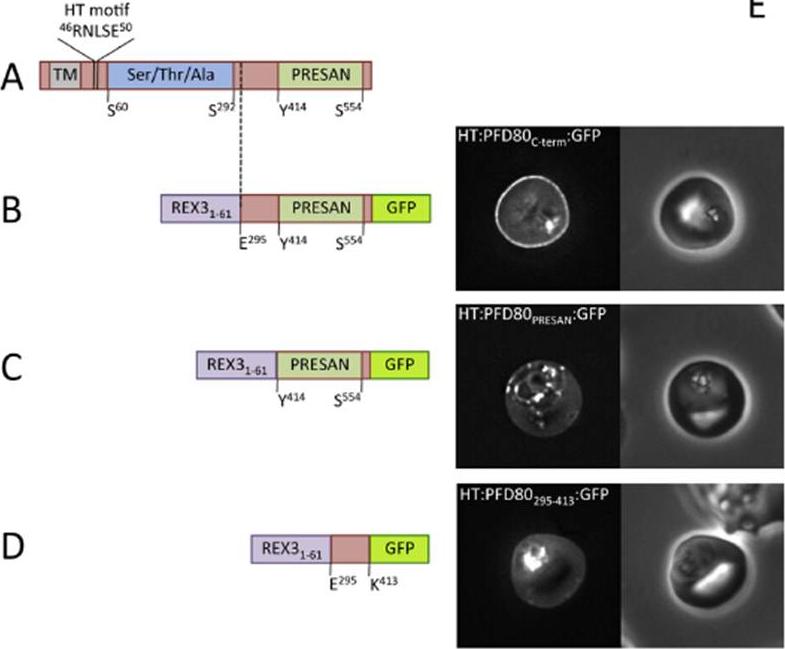
Domain architecture of PF3D7 0401800 (PFD80), not to scale. TM, transmembrane domain/recessed signal sequence; Ser/Thr/Ala refers to serine, threonine andalanine rich-region; PRESAN, domain boundaries defined according to Pfam ID: PF09687. (B–D) Domain architecture and localisation of HT:PFD80:GFP reporter constructs.REX31–61, N-terminal sequence of REX3, including its HT motif, which is sufficient to drive protein export. Residue numbers, below each diagram, refer to position withinPFD80. The left- and right-hand images show GFP localisation and a phase contrast image, respectively. The identity of each parasite line is indicated above the respective images. allowed us to definea novel region sufficient to confer peripheral localisation in thehost erythrocyte. This region of PFD80, comprising both thePRESAN domain and an additional N-terminal flanking sequence,is sufficient to target to the erythrocyte periphery; neither domain alone is functional.Tarr SJ, Moon RW, Hardege I, Osborne AR. A conserved domain targets exported PHISTb family proteins to the periphery of Plasmodium infected erythrocytes. Mol Biochem Parasitol. 2014 196(1):29-40 PMID:
See original on MMP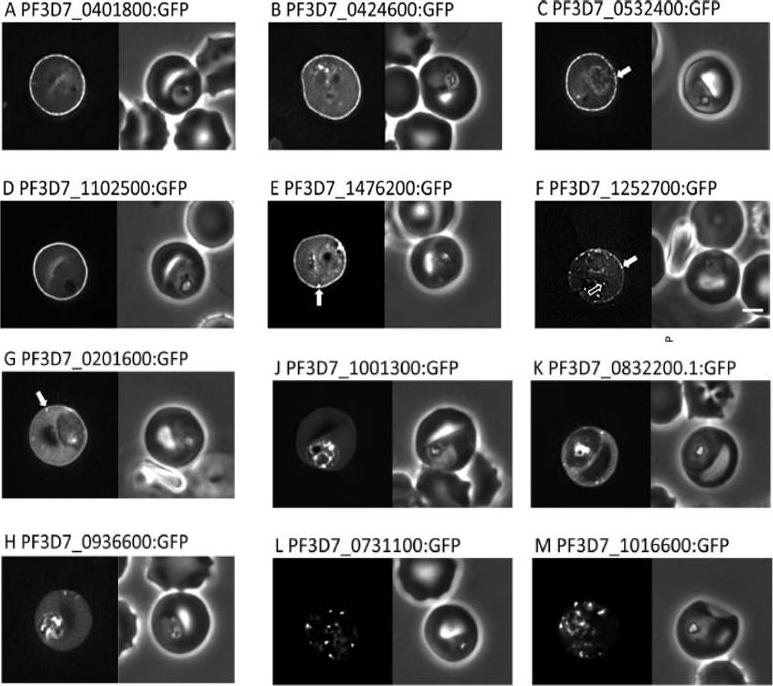
Localisation of PHIST:GFP proteins. The left- and right-hand images show GFP localisation and a phase contrast image, respectively. White filled arrows: peripheral GFP puncta; White unfilled arrow: GFP puncta in host erythocyte cytosol. Scale bar, 2 mm. GFP-tagged PF3D7_0401800, PF3D7_0424600, PF3D7_0532400, PF3D7_1102500 and PF3D7_1476200 were all exported to the host cell and displayed a striking localisation at the edge of the host erythrocyte (1A–E), PF3D7 1252700 was clearly peripheral in the host erythrocyte (F). PF3D7_0201600 (G) shows weak accumulation at the erythrocyte periphery, but the majority of the protein was localised in the RBC cytosol. PF3D7_0936600 was localised in the erythrocyte cytosol, not at the host cell periphery (H).Tarr SJ, Moon RW, Hardege I, Osborne AR. A conserved domain targets exported PHISTb family proteins to the periphery of Plasmodium infected erythrocytes. Mol Biochem Parasitol. 2014 196(1):29-40
See original on MMPMore information
| PlasmoDB | PF3D7_0401800 |
| GeneDB | PF3D7_0401800 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | MAL4P1.16, PFD0080c |
| Orthologs | |
| Google Scholar | Search for all mentions of this gene |