PCHAS_1411800 AP-1 complex subunit mu-1, putative (AP1M1)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
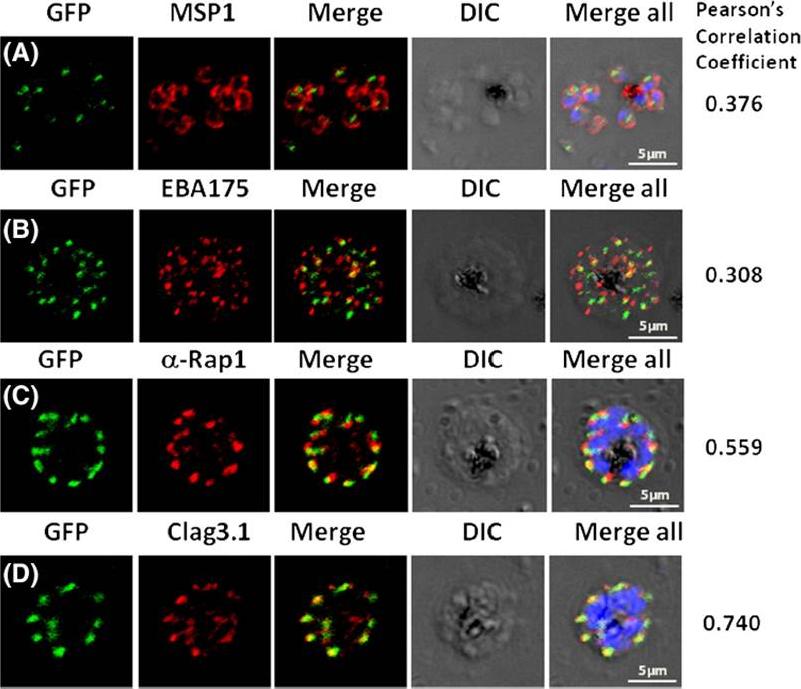
Pfμ1 co-localizeswith rhoptrymarker proteins in schizont stage parasites. Transgenic parasites expressing Pfμ1–GFPwere immunostained with antibodies specific to the Merozoite surface localized MSP1 (A), Microneme localized EBA175 (B), and Rhoptry localized RAP1 (C) and Clag3.1 (D). The parasite nuclei were stained with DAPI and slides were visualized by confocal microscopy. Representative images are shown for each antibody, together with DIC images; scale bars denote 5 μM. To quantify co-localisation, Pearson correlation coefficients of the individual stains were calculated and are shown in the right panel of each image. IFA with antibodies to rhoptry (RAP1 and Clag3.1), microneme (EBA175), and surface markers (MSP1). IFAwith anti-MSP1 antibody showed no overlap in staining betweenMSP1 and Pfμ1 (A). Similar results were seen with antibodies to EBA175 (B). Importantly, anti-RAP1 and anti-Clag3.1 showed co-localization with the Pfμ1–GFP chimeric protein (C and D), suggesting a potential role for Pfμ1 in rhoptry trafficking. Co-localization between Pfμ1 and RAP1 was first observed ~24 h post invasion in budding vesicles near the Golgi. As nuclear division commenced (32 h), Golgi multiplication occurred as well, and this resulted in apical distribution of Pfμ1 along with RAP1 in the rhoptries. Kaderi Kibria KM, Rawat K, Klinger CM, Datta G, Panchal M, Singh S, Iyer GR, Kaur I, Sharma V, Dacks JB, Mohmmed A, Malhotra P. A role for adaptor protein complex 1 in protein targeting to rhoptry organelles in Plasmodium falciparum. Biochim Biophys Acta. 2015 1853(3):699-710.
See original on MMP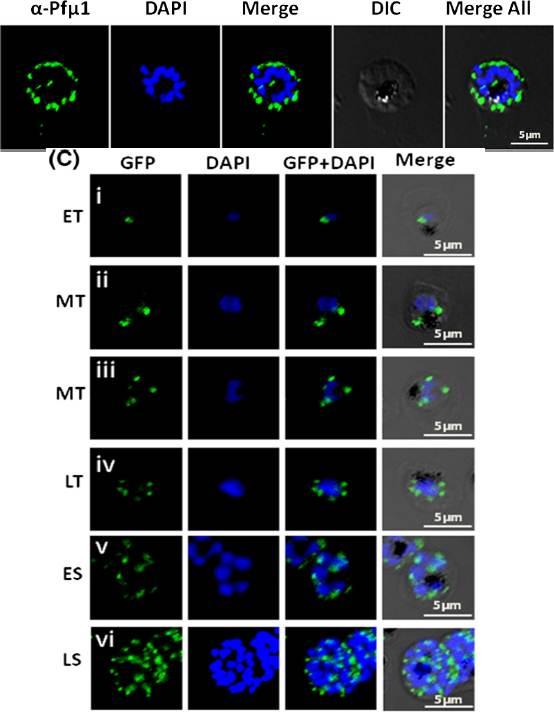
Up: Expression and localization of Pfμ1 protein in P. falciparum. Immunofluorescence analysis using the anti-Pfμ1 antibody, demonstrating discreet punctate structures in schizont stage parasites. Parasite nuclei were stained with DAPI; scale bars denote 5 μm. The well-defined punctate structures in schizont stage of the parasite, a pattern characteristic of staining for apical secretory organelles. Lower panel: Live cell imaging of transgenic parasites expressing the Pfμ1–GFP fusion protein fromearly trophozoite to late schizont stages. Parasite nuclei were stained with DAPI; scale bars denote 5 μM. ET, Early Trophozoite; MT, Mid Trophozoite; LT, Late Trophozoite; ES, Early Schizont; LS, Late Schizont. 10–18 h postinvasion, Pfμ1–GFP was observed as a single spot in a small compartment in close proximity to the nucleus (i). After further development, in young trophozoite stages (20–24 h post invasion), two to four fluorescent puncta were observed adjacent to the nucleus (ii–iv). As nuclear division commences (~32 h post invasion), Pfμ1–GFP was localized in multiple compartments, ensuring that each merozoite inherits one such spot (v). At the mature schizont stage, a welldefined punctate staining was seen, typical of apical organelle distribution of proteins in Plasmodium (vi).Kaderi Kibria KM, Rawat K, Klinger CM, Datta G, Panchal M, Singh S, Iyer GR, Kaur I, Sharma V, Dacks JB, Mohmmed A, Malhotra P. A role for adaptor protein complex 1 in protein targeting to rhoptry organelles in Plasmodium falciparum. Biochim Biophys Acta. 2015 1853(3):699-710.
See original on MMP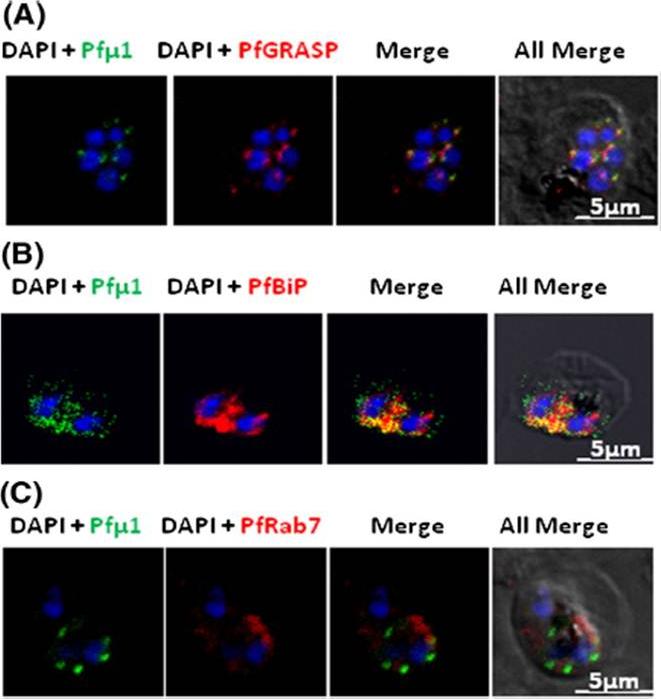
Pfμ1 is closely associated with the Golgi and ER in early trophozoite stages. Transgenic parasites expressing Pfμ1–GFP protein at trophozoite stages were immunostainedwith anti-PfGRASP (A), anti-PfBiP (B) and anti-PfRab7 (C) antibodies. The parasite nuclei were stained with DAPI and slides were visualized by confocal laser scanning microscopy. Scale bars denote 5 μm. Pfμ1 partially co-localized with PfGRASP and PfBip. the ER (which is contiguous with the nuclear envelope) and Golgi in developing merozoites are very closely juxtaposed, and dense vesicular traffic occurs between these two compartments, as well as between nascent rhoptries. No overlap between Pfμ1–GFP and Rab7.This suggests that these proteins do not co-localize, and hence that Rab7 is not involved in trafficking at this stage (3C). These results thus suggest the presence of Pfμ1 in the Golgi–ER network during the early stages of intra-erythrocytic development.Kaderi Kibria KM, Rawat K, Klinger CM, Datta G, Panchal M, Singh S, Iyer GR, Kaur I, Sharma V, Dacks JB, Mohmmed A, Malhotra P. A role for adaptor protein complex 1 in protein targeting to rhoptry organelles in Plasmodium falciparum. Biochim Biophys Acta. 2015 1853(3):699-710.
See original on MMP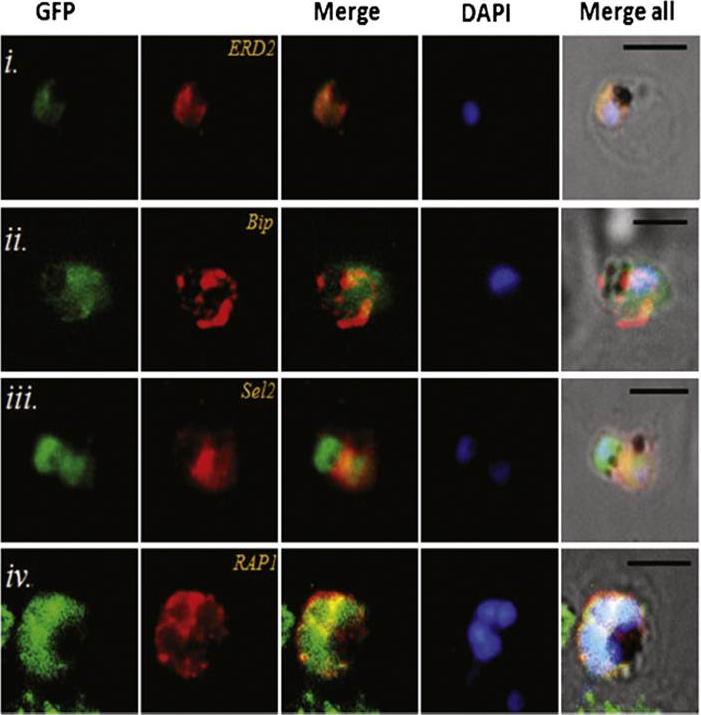
Transgenic parasites expressing Pfμ1–GFP were treated with Brefeldin A (BFA) and immunostained with antibodies specific to cis-Golgi apparatus marker ERD2 (i), endoplasmic reticulum marker, Bip (ii), cytoplasm localized, Sel2 (iii) and RAP1 (iv). The Pfμ1–GFP fusion protein colocalized with Sel2 as well as ERD2 in the parasite cytoplasm upon BFA treatment (i & iii). Parasite nuclei were stained with DAPI; scale bars denote 5 μm. Co-localization with antibodies to ERD2 (a cis-Golgi marker) and BiP, as well as the resident rhoptry protein RAP1 and the cytosolic protein Sel2 showed that Pfμ1 did not show a similar pattern as Bip or Sel2Pfμ1 showed a substantially similar staining pattern as ERD2 which is a Golgi marker (i–iii). Pfμ1 also did co-localize with RAP1 in BFA treated parasites (iv). These observations are consistent with Pfμ1 being Golgi associated at this life stage, consistent with its similar dynamics of redistribution upon brefeldin treatment.Kaderi Kibria KM, Rawat K, Klinger CM, Datta G, Panchal M, Singh S, Iyer GR, Kaur I, Sharma V, Dacks JB, Mohmmed A, Malhotra P. A role for adaptor protein complex 1 in protein targeting to rhoptry organelles in Plasmodium falciparum. Biochim Biophys Acta. 2015 1853(3):699-710.
See original on MMPMore information
| PlasmoDB | PCHAS_1411800 |
| GeneDB | PCHAS_1411800 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PC000656.01.0, PCAS_141180, PCHAS_141180 |
| Orthologs | PBANKA_1409900 , PF3D7_1311400 , PKNH_1411800 , PVP01_1412400 , PVX_122405 , PY17X_1411700 |
| Google Scholar | Search for all mentions of this gene |