PCHAS_0409400 parasite-infected erythrocyte surface protein (PIESP1)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
RMgm-188 | Imported from RMgmDB | |
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
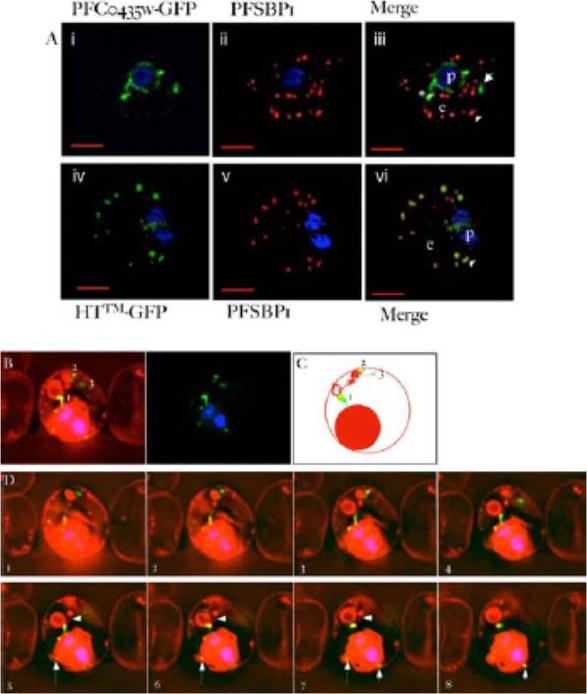
PFC0435w is a junctional protein of the TVN. A) Relative distribution of PFC0435w-GFP (i) or HTTM-GFP (ii) and Maurer’s clefts marked by PfSBP1 (ii, iv) in P. falciparum infected erythrocytes.The cells were also stained with Hoechst 33342 (blue) to visualize the DNA of the parasite. Panels i, ii and iv, v are single optical sections, whose merge is shown in iii and vi respectively. Scale bar, 2 mm. B–D) Association of PFC0435w-GFP with membrane buds and TVN structures B. Zero degree projection of an erythrocyte infected with a PFC0435w- GFP-expressing parasite stained with Rhodamine B. The numbers indicate the large circular TVN structure (1), and PFC0435w-GFP associated with other parts of the TVN (2 and 3). C) Cartoon representation of the infected cell in panel B. D) Optical sectioning of the infected erythrocyte shown in panel B. Distance between sections is 0.2 mm. Note the progression of PFC0435w-GFP staining from the PVM (sections 1–4) to the circular Rhodamine B-stained structure in the erythrocyte (sections 4–8). Arrows indicate PVM buds colocalizing with PFC0435w-GFP. Arrowhead indicates TVN connections between two large loops.van Ooij C, Tamez P, Bhattacharjee S, Hiller NL, Harrison T, Liolios K, Kooij T, Ramesar J, Balu B, Adams J, Waters AP, Janse CJ, Haldar K. The malaria secretome: from algorithms to essential function in blood stage infection. PLoS Pathog. 2008 Jun 13;4(6):e1000084,
See original on MMP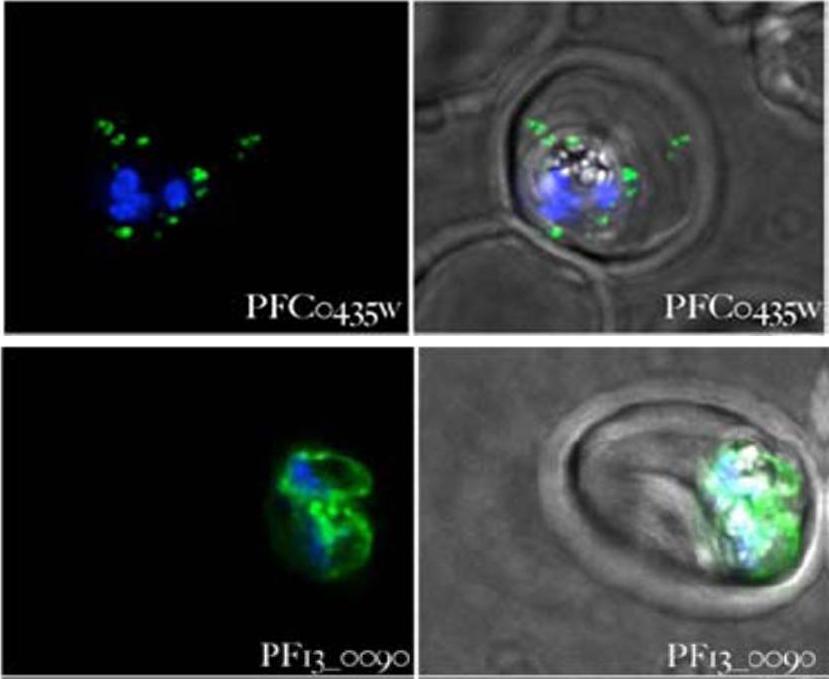
Parasites expressing the gene indicated in each panel fused to gfp were stained with Hoechst 33342 and examined 18–36 hours after infection. Panels on the left-hand side show the overlap of the Hoechst 33342 staining and the GFP fluorescence, the panels on the right show the Hoechst 33342 staining, the GFP fluorescence and a phase contrast image of the infected cell. All images are composites of multiple consecutive optical sections. PFC0435w is seen exported to the erythrocyte. It has a transmembrane spanning domain, it displays punctate spots in the erythrocyte as well as closely associated with the PV. PF13_0090 is not detected within the erythrocyte and probably not exported.van Ooij C, Tamez P, Bhattacharjee S, Hiller NL, Harrison T, Liolios K, Kooij T, Ramesar J, Balu B, Adams J, Waters AP, Janse CJ, Haldar K. The malaria secretome: from algorithms to essential function in blood stage infection. PLoS Pathog. 2008 Jun 13;4(6):e1000084, PMID
See original on MMP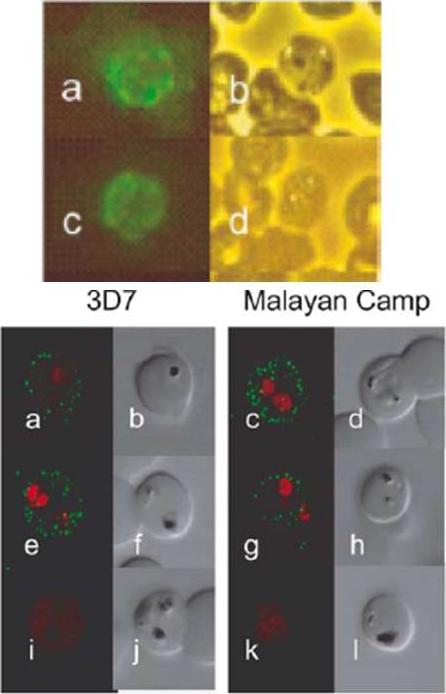
Localization of PIESP1 and PIESP2 with specific antibodies. (A) Immunofluorescent microscopy of PIEs (P. falciparum strain 3D7) fixed onto glass slides. Glass slide were incubated with anti-PIESP1 (Plates a and b), anti-PIESP2 (Plates c and d), and pooled preimmune sera (Plates e and f). Fluorescent images (Plates a, c, and e) and phase-contrast images (Plates b, d, and f) were collected simultaneously on an Olympus BX-60. (B) Localization of PIESPs with immunofluorescent confocal microscopy. Live P. falciparum strains 3D7 and Malayan Camp (unselected for knob and rosetting phenotypes) were incubated in suspension with specified antibodies. Plates a–d, anti-PIESP1 as primary antibody; Plates e–h, anti-PIESP2 as primary antibody; Plates i–l, preimmune sera as primary antibody. The 488 and 568 nm laser channels, for Alexa Fluor 488 and ethidine bromide, respectively, were collected separately and later superimposed (Plates a, c, e, g, i, and k). The DIC images (Plates b, d, f, h, j, and l) were collected simultaneously using the transmitted light detector. Florens L, Liu X, Wang Y, Yang S, Schwartz O, Peglar M, Carucci DJ, Yates JR 3rd, Wub Y. Proteomics approach reveals novel proteins on the surface of malaria-infected erythrocytes. Mol Biochem Parasitol. 2004 135:1-11. Copyright Elsevier 2010.
See original on MMP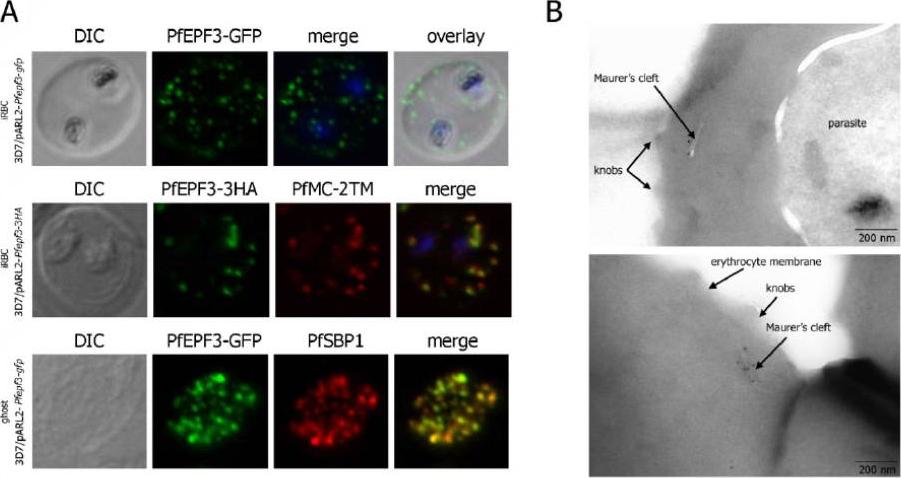
Localization and topology of the PfEPF3 proteins. A. Fluorescent patterns of iRBCs infected by 3D7/pARL2-Pfepf3-gfp (live imaging) and 3D7/pARL2-Pfepf3-3HA (immunodetection using anti-HA antibodies, in green, and anti-PfMC-2TM antibodies, in red) and of resealed ghosts from 3D7/pARL2-Pfepf3-gfp iRBCs (GFP fluorescence, in green, and immunodetection using anti-SBP1 antibodies, in red). RBCs and ghost preparations were incubated with DAPI for nucleus labelling. B. Immunoelectron microscopy using anti-GFP antibodies.Mbengue A, Audiger N, Vialla E, Dubremetz JF, Braun-Breton C. Novel Plasmodium falciparum Maurer's clefts protein families implicated in the release of infectious merozoites. Mol Microbiol. 2013 88(2):425-42 PMID: .
See original on MMPMore information
| PlasmoDB | PCHAS_0409400 |
| GeneDB | PCHAS_0409400 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PC001354.02.0, PC300113.00.0, PC400567.00.0, PCAS_040940, PCHAS_040940 |
| Orthologs | PBANKA_0408500 , PF3D7_0310400 , PKNH_0832300 , PVP01_0829800 , PVX_119625 , PY17X_0411000 |
| Google Scholar | Search for all mentions of this gene |