PCHAS_0109400 histone H3, putative
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
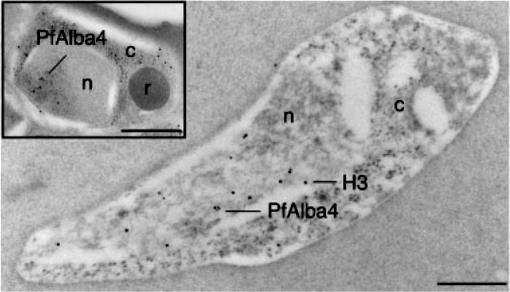
Immuno-EM was performed on ring stage parasites using anti-PfAlba4 antibodies (Ab 237–2) bound to 10nm gold particles and anti-H3 antibodies bound to 15nM gold particles. The inset shows a single developing merozoite within schizont-stage parasites labeled with anti-PfAlba4 antibodies (Ab 237–2) bound to 10nm gold particles; n=nucleus, c=cytoplasm, r=rhoptry; Scale bars represent 500 nm. In the ring stage, we detected PfAlba4 uniquely in the nucleus. Since the nuclear membrane is difficult to visualize in immuno-EM in the ring stage, co-labeling with antibodies directed against histone H3 was used to determine the area of the nucleus in this parasite form. The images localize PfAlba4 at the nuclear periphery. This pattern was confirmed in developing merozoites (inset) where distinct clusters of PfAlba4 could be detected at the periphery of the nuclei.Chêne A, Vembar SS, Rivière L, Lopez-Rubio JJ, Claes A, Siegel TN, Sakamoto H, Scheidig-Benatar C, Hernandez-Rivas R, Scherf A. PfAlbas constitute a new eukaryotic DNA/RNA-binding protein family in malaria parasites. Nucleic Acids Res. 2011 Dec 13. [Epub ahead of print]
See original on MMP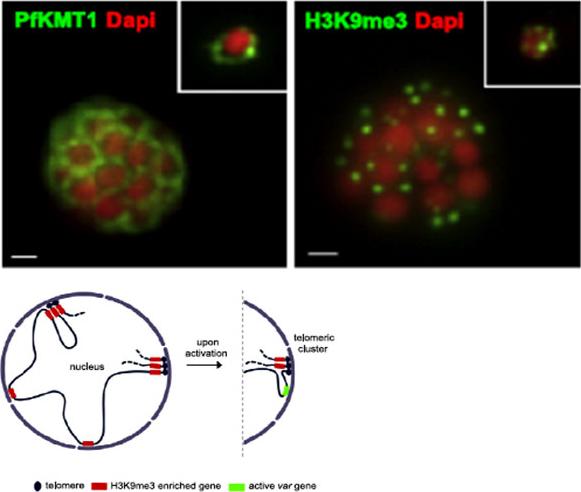
PfKMT1 and genes enriched in H3K9me3 are positioned at the nuclear periphery. Immuno-fluorescence assays using antibodies against H3K9me3 H3 (right) and the P. falciparum putative H3K9 methyltransferase PfKMT1 (left). 3D7 parasites are in late schizont (main image) and ring stage (top-right square). Nuclear DNA was detected with DAPI (red).Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009 5:179-90.
See original on MMP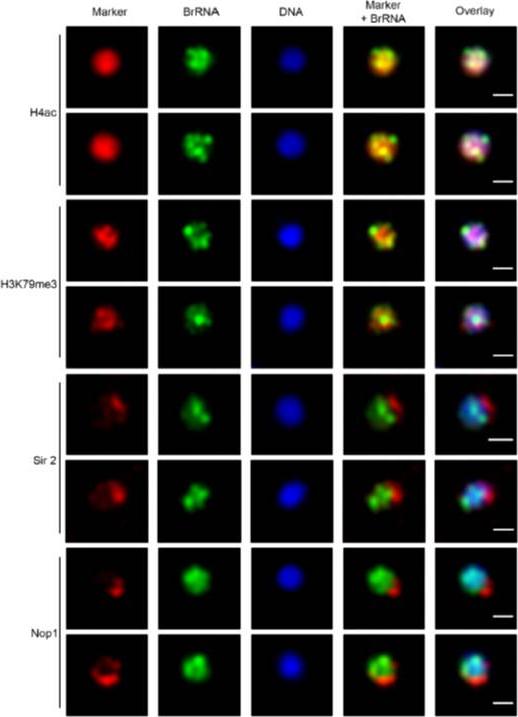
Transcription sites form a distinct compartment in the nucleus of P. falciparum. Immunofluorescence assay for nascent RNAs of ring stage parasites labeled with BrUTP (BrRNA, green) and the compartments of acetylated H4 (H4ac, active chromatin); a proposed histone marker of nascent transcription (H3K79me3; PfSir2A (silencing compartment); and Nop1 (nucleolar compartment). DNA was labeled by DAPI, in blue. Bars, 1 mm.Moraes CB, Dorval T, Contreras-Dominguez M, Dossin Fde M, Hansen MA, Genovesio A, Freitas-Junior LH. Transcription sites are developmentally regulated during the asexual cycle of Plasmodium falciparum. PLoS One. 2013;8(2):e55539.
See original on MMP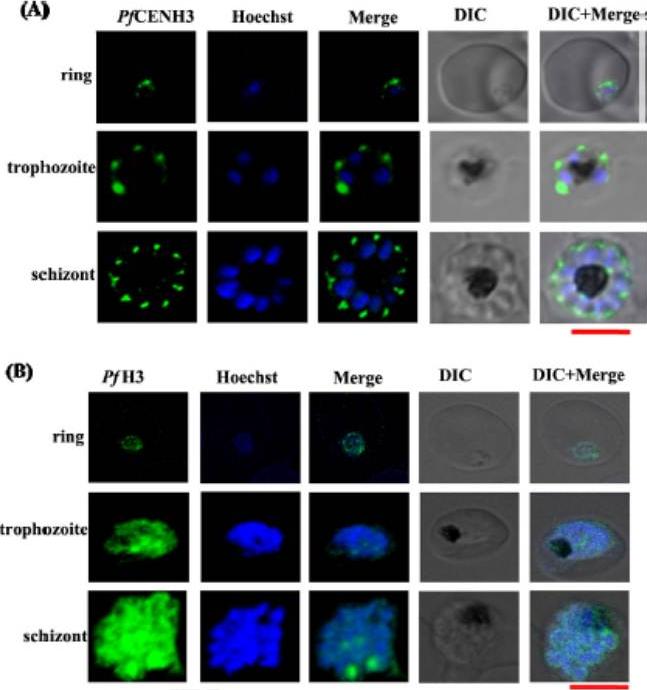
PfCENH3 is expressed at the perinuclear regions while PfH3 localizes to nucleus. (A, B) The confocal images for immuno-fluorescence assays represent the subcellular localization pattern of PfCENH3 and PfH3 (green) with respect to the Hoechst stained nucleus (blue) at ring, trophozoite and schizont stages of P. falciparum respectively. While PfH3 localizes throughout the nuclear mass, PfCENH3 localizes towards perinuclear regions at various intraerythrocytic stages. At ring stage, PfCENH3 localizes as a single cluster per nucleus. At trophozoite and schizont stages it localizes as discrete spots on each nucleus. Scale bar = 5 mm.Verma G, Surolia N. Plasmodium falciparum CENH3 is able to functionally complement Cse4p and its, C-terminus is essential for centromere function. Mol Biochem Parasitol. 2013 Dec 6. [Epub ahead of print]
See original on MMP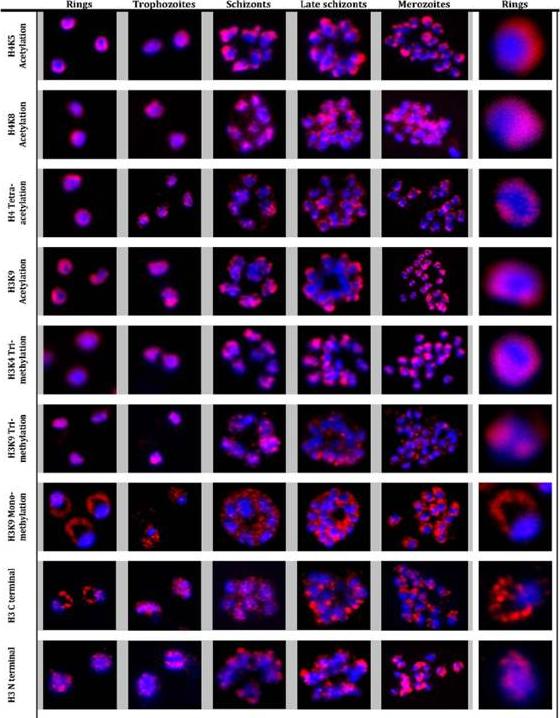
Immunofluorescence analysis of histone modifications in P. falciparum. Localization of histone modifications were analyzed with the ring, trophozoite, schizont, late schizont and merozoite stages of the IDC. IFAs were carried out with antibodies against specific histone 3 and 4 lysine residue acetylations: H3K9Ac, H4K5Ac, H4K8Ac, and H4Ac4, as well as methylations: H3K4Me3, H3K9Me3 and H3K9Me1, and unmodified histone H3. Nuclear DNA was stained with DAPI (blue). All modifications, with the exception of H3K9Me1 and H3 (antibody raised against H3 C terminal), showed specific and distinct localization in the nucleus in all the stages. In contrast, H3K9Me1 was localized mainly outside the nucleus with very low levels detected inside the nucleus. Luah YH, Chaal BK, Ong EZ, Bozdech Z. A moonlighting function of Plasmodium falciparum histone 3, mono-methylated at lysine 9? PLoS One. 2010 5(4):e10252.
See original on MMP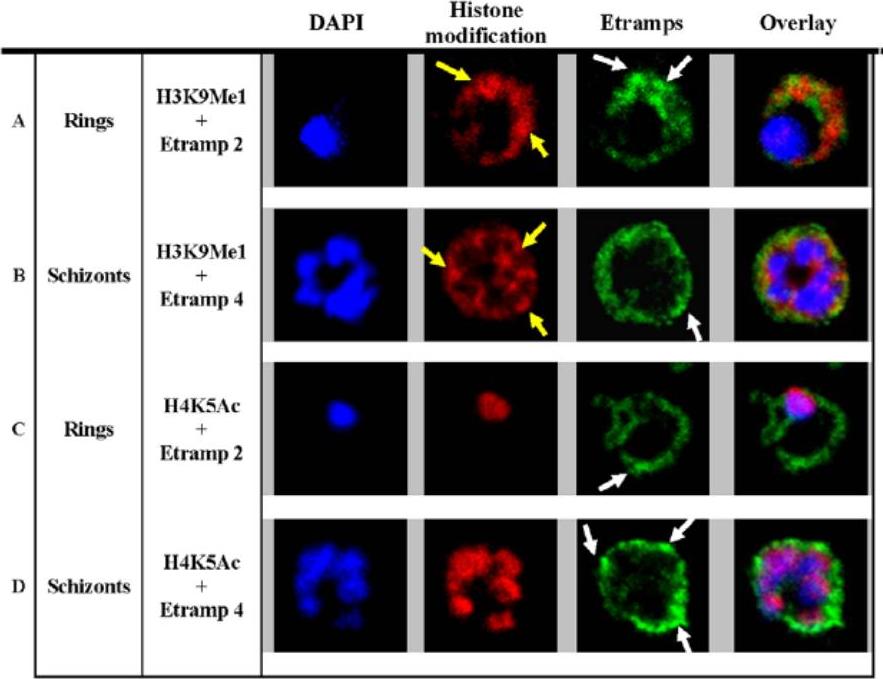
H3K9Me1 localized to the parasitophorous vacuole during the ring stage. Co-localization of H3K9Me1 with Etramp 2 (A) and 4 (B) was performed in ring and schizont stage parasites respectively. Similarly, co-localization of H4K5Ac with Etramp 2 (C) and 4 (D) was performed in ring and schizont stage parasites respectively. H3K9Me1/H4K5Ac and Etramp 2/4 were stained red and green respectively. DAPI stained nuclear DNA blue. Yellow and white arrows indicate foci of more intense fluorescence produced by H3K9Me1 and Etramp labeling respectively. In ring stage parasites, compared to schizonts, H3K9Me1 partially co-localized with Etramp 2 indicating localization to different compartments of the PV. H4K5Ac was localized solely to the nucleus and did not co-localize with either Etramp 2 or 4.Luah YH, Chaal BK, Ong EZ, Bozdech Z. A moonlighting function of Plasmodium falciparum histone 3, mono-methylated at lysine 9? PLoS One. 2010 5(4):e10252.
See original on MMP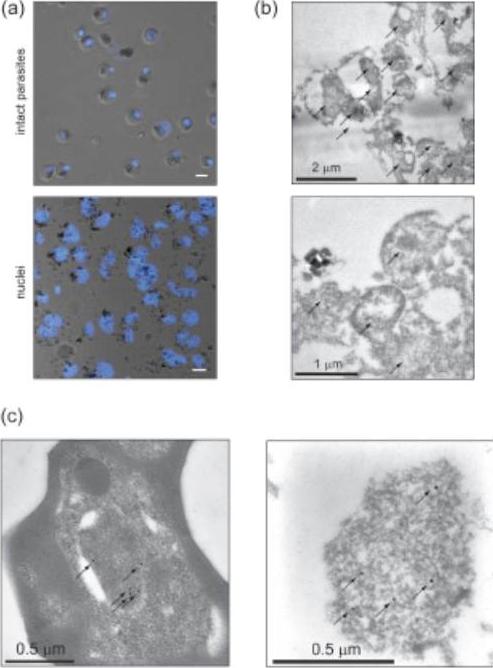
Preparation of crude P. falciparum nuclei for proteomic analysis. (a) Visual assessment of the crude nuclear preparation. Intact parasites (control) and isolated nuclei were stained with DAPI and analysed by DIC and fluorescence microscopy. Scale bar 2 mm. (b) TEM analysis of isolated nuclei shows intact as well as damaged nuclei and very little contamination from other organelles. Arrows indicate individual nuclei. (c) Immunoelectron microscopy verifies the identity of nuclei. Anti-H3K4me3 antibodies were used to label nuclei in intact cells (left panel) and in the nuclear preparation (right panel) (arrows). Oehring SC, Woodcroft BJ, Moes S, Wetzel J, Dietz O, Pulfer A, Dekiwadia C, Maeser P, Flueck C, Witmer K, Brancucci NM, Niederwieser I, Jenoe P, Ralph SA, Voss TS. Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol. 2012 Nov 26;13(11):R108.
See original on MMP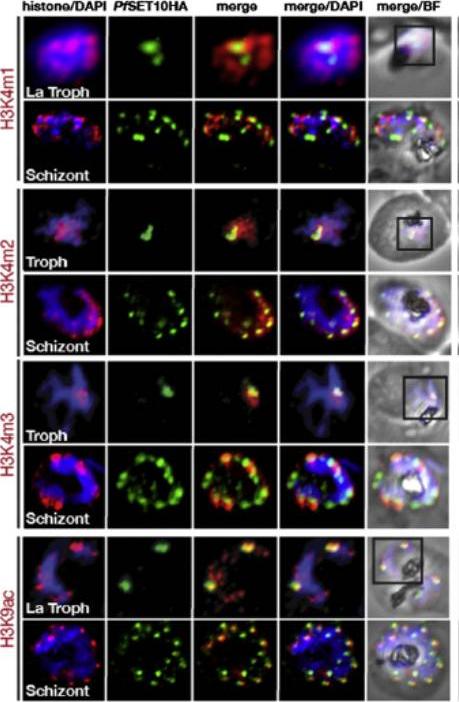
Colocalization of histone H3 marks with PfSET10. Shown are from top to bottom: H3K4m1, H3K4m2, H3K4m3, and H3K9ac. The histone marks (red) were detected using specific antibodies and colocalized with PfSET10-HA (green) using anti-HA antibodies. Nucleus position is shown by DAPI staining. PfSET10 colocalized with the H3K4me1, me2, and me3 marks in the perinuclear region, which is also marked by H3K9ac Volz JC, Bártfai R, Petter M, Langer C, Josling GA, Tsuboi T, Schwach F, Baum J, Rayner JC, Stunnenberg HG, Duffy MF, Cowman AF. PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe. 2012 11(1):7-18.
See original on MMP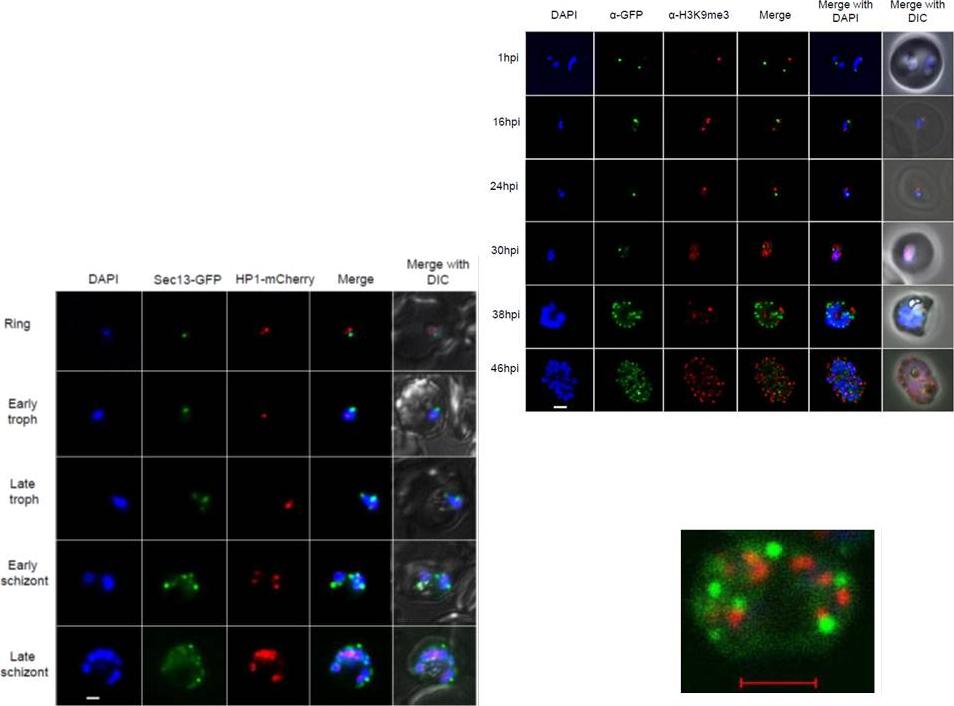
Right: PfSec13 does not co-localize with P. falciparum heterochromatin. IFAs of the PfSec13-GFP expressing parasites that were co-immunolabled with α-GFP (green) and α-H3K9me3 (red) at different stages of the IDC. In 85% of the counted nuclei of all stages (n=100), the H3K9me3 signal and PfSec13-GFP did not co-localize.Left: In vivo imaging of parasites expressing both PfSec13-GFP and PfHP1-HA-mCherry during the IDC. Although PfSec13 (green) and PfHP1 (red) localized to the nuclear periphery they do not co-localize.Righet lower corner: In vivo confocal microscopy of parasites expressing both PfSec13-GFP and PfHP1-HA-mCherry.Dahan-Pasternak N, Nasereddin A, Kolevzon N, Pe'er M, Wong W, Shinder V, Turnbull L, Whitchurch CB, Elbaum M, Gilberger TW, Yavin E, Baum J, Dzikowski R. PfSec13 is an unusual chromatin associated nucleoporin of Plasmodium falciparum, which is essential for parasite proliferation in human erythrocytes. J Cell Sci. 2013 126(Pt 14):3055-69 Reproduced with permission
See original on MMP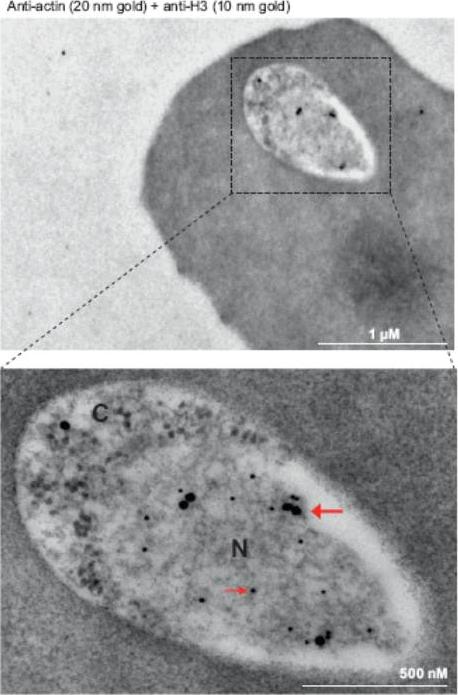
Immunoelectron microscopy of 3D7 rings. Double labeling with mouse anti-Pfactin-I (20 nm gold grain, bold arrow) and rabbit anti-histone H3 (10 nm gold grain, slim arrow). N, nucleus; C, cytoplasm. Gold grains are virtually confined to the nuclear periphery.Zhang Q, Huang Y, Zhang Y, Fang X, Claes A, Duchateau M, Namane A, Lopez-Rubio JJ, Pan W, Scherf A. A critical role of perinuclear filamentous actin in spatial repositioning and mutually exclusive expression of virulence genes in malaria parasites. Cell Host Microbe. 2011 10:451-63.
See original on MMP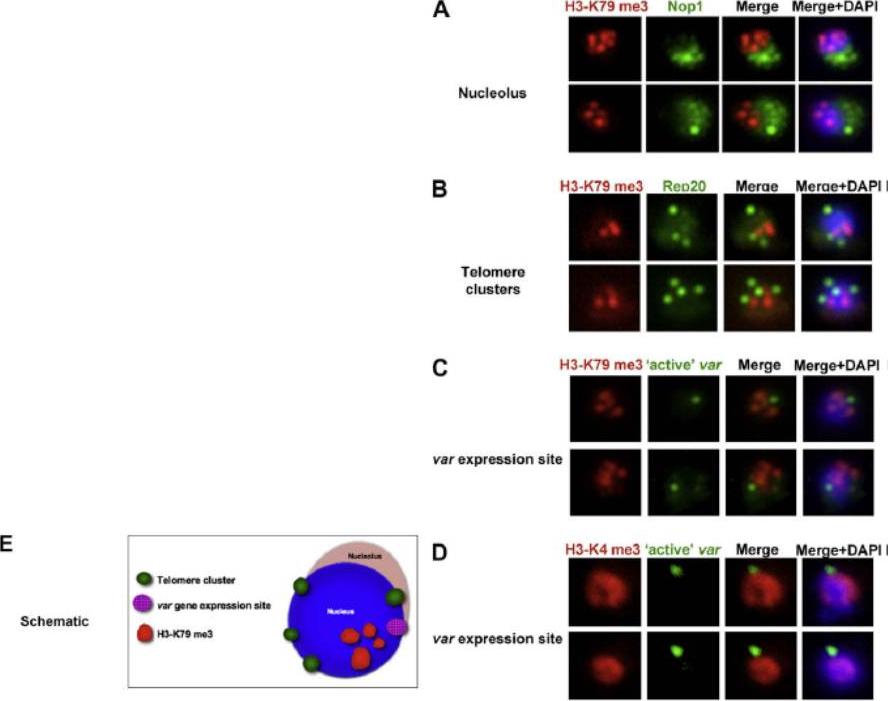
Co-localisation of H3-K79me3 with known cellular compartments. A. Nucleolus: Double labelling IFAs with antibodies against H3-K79me3 and nucleolus marker, PfNop1. B. Telomere clusters: IFeFISH analysis between H3-K79me3 (red) and Rep20 (green). C. Active var gene: IF-FISH analysis between H3-K79me3 (red) and active var gene (green). D. Active var gene colocalisation with activating mark, H3-K4me3. IF-FISH analysis between H3-K4me3 (red) and active var gene (green). DAPI stained nucleus in blue. Font colour represents corresponding fluorescein labelled antibody. Scale bar depicts 1 mm. E. Schematic of the dynamic compartmen-talisation of the P. falciparum nucleus. The distinct sub-nuclear distribution of histone methylation modification, taking example of H3-K79me3 as observed in this study. Known compartments compatible with active transcription are indicated. H3-K79me3 occupies a distinct zone.Issar N, Ralph SA, Mancio-Silva L, Keeling C, Scherf A. Differential sub-nuclear localisation of repressive and activating histone methyl modifications in P. falciparum. Microbes Infect. 2009 11:403-7.
See original on MMP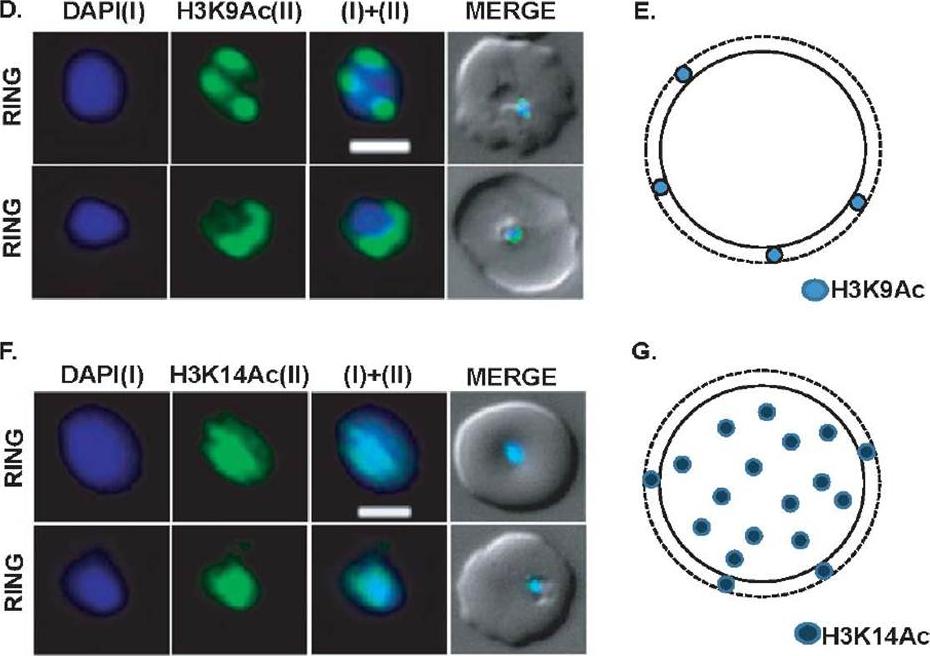
Expression and sub-cellular localization patterns of H3K9Ac and H3K14Ac histone marks. (D) Perinuclear localization pattern of acetylated H3K9Ac marks. Immunofluorescence analysis using antibodies specific for H3K9Ac confirms horseshoe-shaped pattern around nuclear periphery. H3K9Ac also localizes as sharp 3-7 perinuclear foci in majority of the parasites. (E) Schematic representation of H3K9Ac mark in parasite nuclei. (F) Localization pattern of H3K14Ac mark in the parasite nuclei. Unlike H3K9Ac, H3K14Ac signal shows even distribution on the DAPI. (G) Schematic diagram of H3K14Ac marks in the parasite nuclei. Scale bar measures 1 μM. DAPI stains ring nuclei.Srivastava S, Bhowmick K, Chatterjee S, Basha J, Kundu TK, Dhar SK. Histone H3K9 acetylation level modulates gene expression and may affect parasite growth in human malaria parasite Plasmodium falciparum. FEBS J. 2014 Sep 22. [Epub ahead of print]
See original on MMP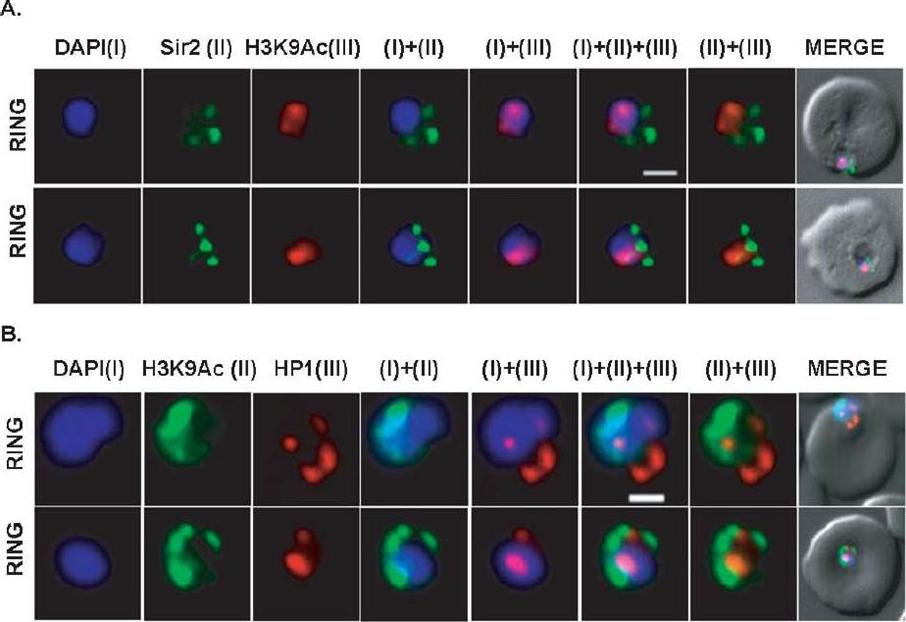
Differential localization patterns of H3K9Ac, PfSir2 and PfHP1. (A) Differential distribution of PfSir2 and H3K9Ac in Plasmodium falciparum. Dual Immuno-fluorescence analysis of PfSir2 (green) and H3K9Ac (red) reveals PfSir2 as sharp foci around nuclei and H3K9Ac as sharp foci closely attached to nuclei periphery. There is no colocalization of PfSir2 and H3K9Ac in ring stage. Scale bar measures 1 μM. DAPI stains nuclei. (B) Differential distribution of H3K9Ac (green) and PfHP1, specific for H3K9me3 repressive histone marks (red). Dual immuno-fluorescence assay shows no colocalization between these two proteins. Scale bar measures 1 μM. DAPI stains nuclei.Srivastava S, Bhowmick K, Chatterjee S, Basha J, Kundu TK, Dhar SK. Histone H3K9 acetylation level modulates gene expression and may affect parasite growth in human malaria parasite Plasmodium falciparum. FEBS J. 2014 Sep 22. [Epub ahead of print]
See original on MMP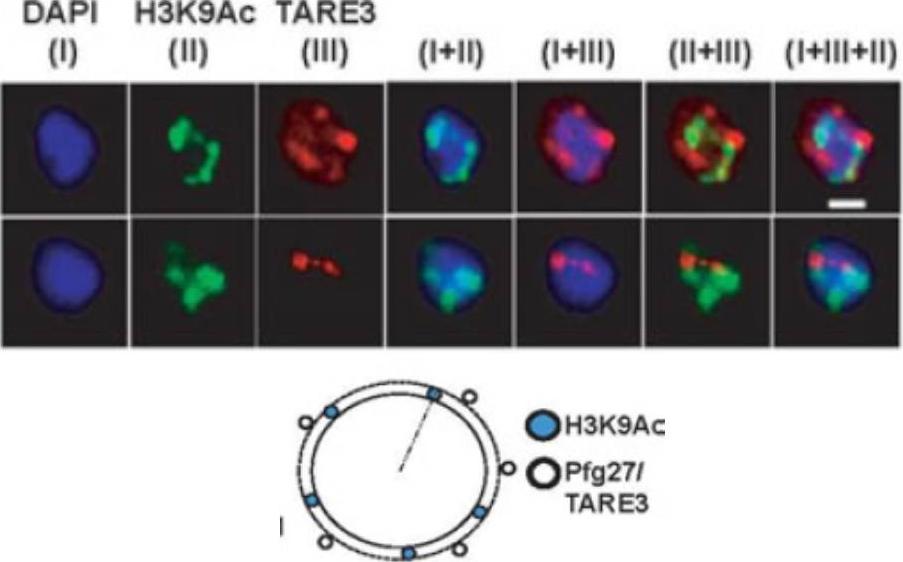
Active promoter localizes to H3K9Ac enriched zones located at nuclear periphery. No colocalization takes place between TARE3 sub-telomeric regions and H3K9Ac marks. FISH-IFA experiments were performed using labeled TARE3 (red) DNA probe and antibodies specific for H3K9Ac (green) marks. Left scheme: Schematic representation of differential localizations of TARE3 and H3K9Ac marks. DAPI stains ring nuclei. Scale bar measures 1 μM.Srivastava S, Bhowmick K, Chatterjee S, Basha J, Kundu TK, Dhar SK. Histone H3K9 acetylation level modulates gene expression and may affect parasite growth in human malaria parasite Plasmodium falciparum. FEBS J. 2014 Sep 22. [Epub ahead of print]
See original on MMPMore information
| PlasmoDB | PCHAS_0109400 |
| GeneDB | PCHAS_0109400 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PCHAS_010940 |
| Orthologs | PBANKA_0108800 , PF3D7_0610400 , PKNH_1139900 , PVP01_1138700 , PVX_113665 , PY17X_0110400 |
| Google Scholar | Search for all mentions of this gene |