PBANKA_1410300 M1-family alanyl aminopeptidase, putative (M1AAP)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
RMgm-806 | Imported from RMgmDB | |
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
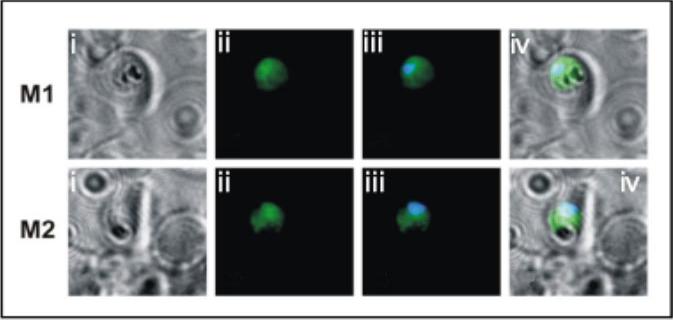
Indirect immunofluorescence of transgenic parasites stained with monoclonal anti c-myc primary antibody followed by anti-mouse cy2. (i) Bright field; (ii) anti-c-myc antibody; (iii) anti-c-myc/nuclear stain merged; (iv) merge of i and iii. The data show that the PfA-M1 transgenic protein is localized to the parasite cytosol. McGowan S, Porter CJ, Lowther J, Stack CM, Golding SJ, Skinner-Adams TS, Trenholme KR, Teuscher F, Donnelly SM, Grembecka J, Mucha A, Kafarski P, Degori R, Buckle AM, Gardiner DL, Whisstock JC, Dalton JP. Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase. Proc Natl Acad Sci U S A. 2009 106:2537-42.
See original on MMP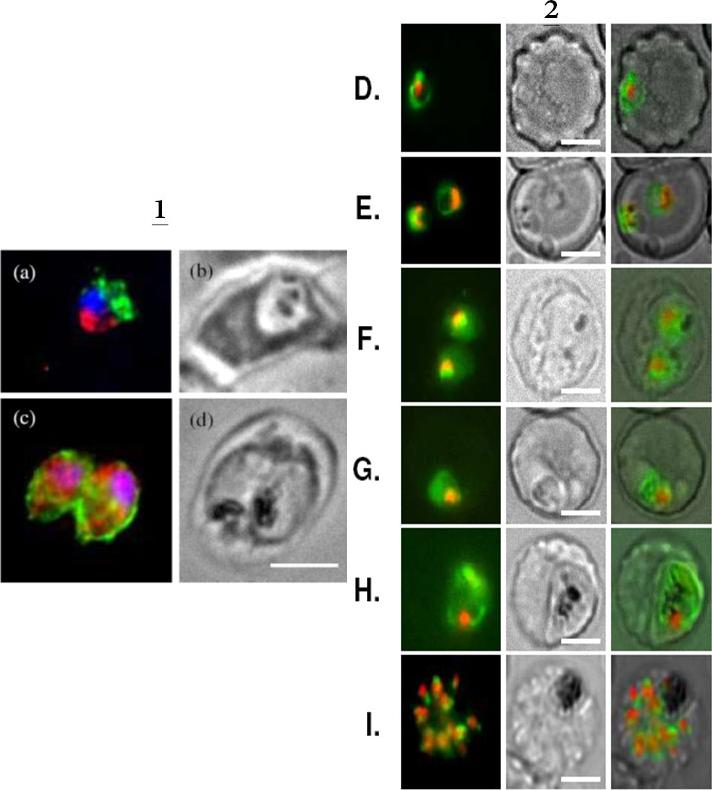
1. BFA-treated (a) and control parasites in presence of ethanol (c) were analysed by immunofluorescence by using anti-MAP1 antibodies to detect PfA-M1 (red) and a mouse anti-exp2 serum as positive control (green). Nuclei, stained by using Hoechst 33342 appear in blue Corresponding phase contrasts are in (b, d). Scale bar, 5μm. In the control exp-2 was targeted as expected to the PVM and PfA-M1 was diffuse in the parasite (but outside the FV)2. Live imaging of FcB1 parasites transfected with a construct allowing expression of PfA-M1-GFP chimera confirm its marginal association with the food vacuole. Live imaging of transfected parasites showing: D, E. ring stages. F, G, trophozoite stages; H. young schizont. I. segmenting schizont. First column: GFP (green) and nuclei (Hoechst 33342); second column: phase contrast; third column: overlay. Scale bar, 5μm.Azimzadeh O, Sow C, Geze M, Nyalwidhe J, Florent I. Plasmodium falciparum PfA-M1 aminopeptidase is trafficked via the parasitophorous vacuole and marginally delivered to the food vacuole. Malar J. 2010 9:189:9
See original on MMP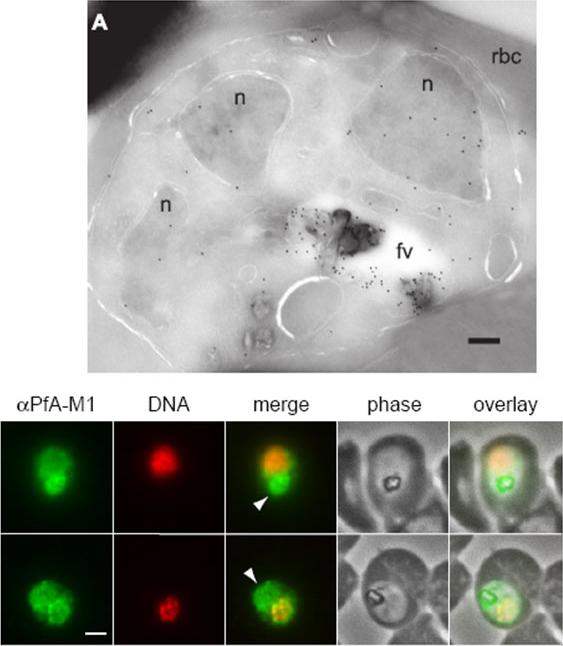
Upper: Immunofluorescence localization of PfA-M1 in parasites fixed with 50% methanol/50% ethanol at -20 °C. Anti-PfA-M1 fluorescence is pseudocolored green and DAPI fluorescence (DNA) is pseudocolored red. Food vacuole fluorescence is identified by co-localization with the hemozoin crystal and is indicated with an arrowhead in the “merge” image. Panel B clearly shows PfA-M1 label in the food vacuole, whereas a nuclear localization is more evident in panel C. Scale bar, 1 μm.Lower: Localization of PfA-M1 in intraerythrocytic P. falciparum. A, PfA-M1 was detected in a parasite cryosection with affinity-purified anti-PfA-M1 antibodies. PfA-M1 is present throughout the lumen of the food vacuole and in the nucleus. fv, food vacuole; n, nucleus; rbc, red blood cell. Scale bar, 250 nmRagheb D, Dalal S, Bompiani KM, Ray WK, Klemba M. Distribution and Biochemical Properties of an M1-family aminopeptidase in Plasmodium falciparum indicate a role in vacuolar hemoglobin catabolism. J Biol Chem. 2011 286:27255-65
See original on MMP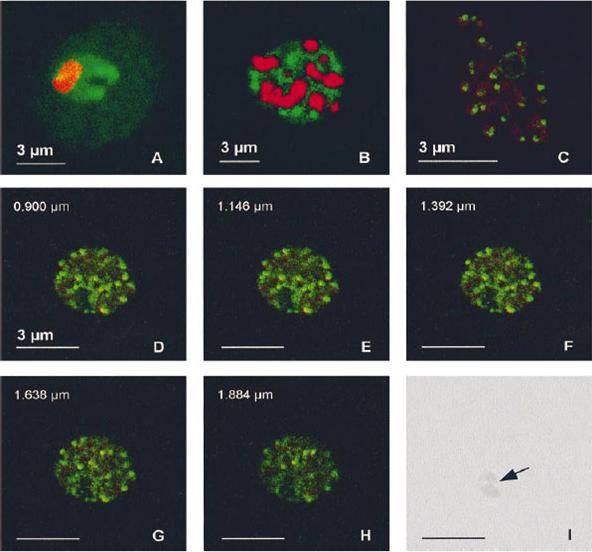
PfA-M1 subcellular location. Samples from Plasmodium falciparum in vitro cultures at various stages were fixed in a mix of methanol and acetone (1:4) and the fuorescence associated with the anti-MAP1 Igs (10 mg/ml, green) and with the nuclei (red) was observed by confocal microscopy. (A) Trophozoite. (B) Schizont. (C) Released merozoites. (D-H) Series of 5 optical sections through a mature schizont (positions along the z axis indicated on the upper left of each panel). (I) Corresponding phase-contrast image, showing the digestive vacuole (indicated by an arrow). In trophozoites, the labelling was diffuse in the parasite cytoplasm, with accumulations around the food vacuole. In schizonts, it turned progressively to a vesicle-like pattern, ending as a clear spot in released merozoites.Allary M, Schrevel J, Florent I. Properties, stage-dependent expression and localization of Plasmodium falciparum M1 family zinc-aminopeptidase. Parasitology. 2002 125:1-10. Copyright Cambridge University Press.
See original on MMP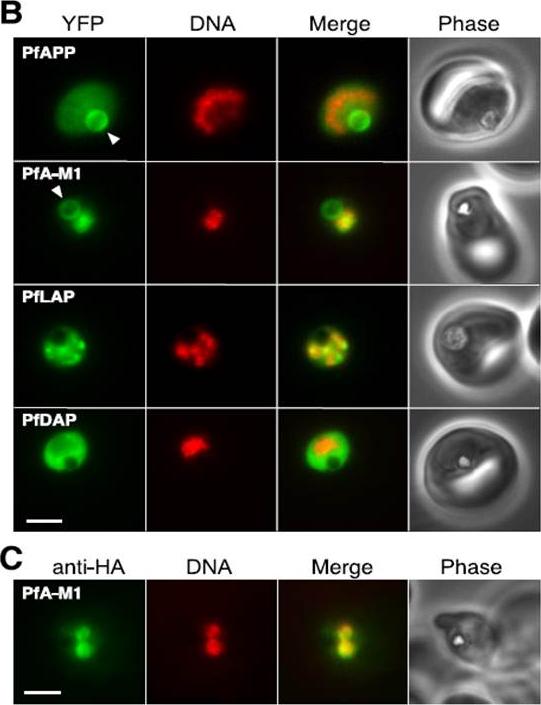
Localization of aminopeptidases in P. falciparum blood stages.PfAPP Aminopeptidase P; PfA-M1 Aminopeptidase N; PfLAP Leucyl aminopeptidase; PfDAP Aspartyl aminopeptidase B, Epifluorescence microscopy of live parasites expressing fusions of YFP with the indicated aminopeptidases. YFP fluorescence is pseudocolored green and Hoechst 33342 (DNA) fluorescence is pseudocolored red. The arrowhead indicates the food vacuole. Bar, 2 mm. C, indirect immunofluorescence detection of PfA-M1-HA. Secondary antibody fluorescence is pseudocolored green and DAPI (DNA) fluorescence is pseudocolored red. Bar, 2 mm.Dalal S, Klemba M. Roles for two aminopeptidases in vacuolar hemoglobin catabolism in Plasmodium falciparum. J Biol Chem. 2007 282:35978-87.
See original on MMP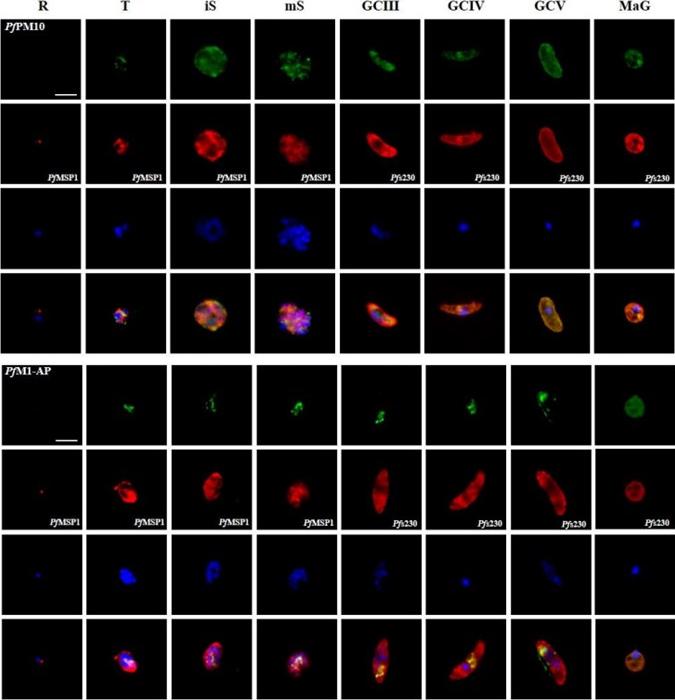
Protein expression profile of selected P. falciparum proteases. IFAs using polyclonal mouse antisera directed against 12 of the proteases was used to demonstrate protein expression in the asexual blood stages (R, ring; T, trophozoite; iS, immature schizont; mS, mature schizont) and gametocytes (GC stages III, IV and V) as well as in macrogametes (MaG) at 20 min post-activation (in green). The asexual and the sexual blood sstages were visualized via immunolabelling with polyclonal rabbit antisera against PfMSP1 and Pfs230, respectively (in red). Hoechst 33342 staining was used to highlight the parasite nuclei (in blue). Bar, 5 μm. Results are representative of three to four independent experiments.Weißbach T, Golzmann A, Bennink S, Pradel G, Julius Ngwa C. Transcript and protein expression analysis of proteases in the blood stages of Plasmodium falciparum. Exp Parasitol. 2017 Mar 25. [Epub ahead of print]
See original on MMPMore information
| PlasmoDB | PBANKA_1410300 |
| GeneDB | PBANKA_1410300 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PB000843.02.0, PBANKA_141030 |
| Orthologs | PCHAS_1412200 , PF3D7_1311800 , PKNH_1412500 , PVP01_1412800 , PVX_122425 , PY17X_1412100 |
| Google Scholar | Search for all mentions of this gene |