PBANKA_1347700 isoleucine--tRNA ligase, putative
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
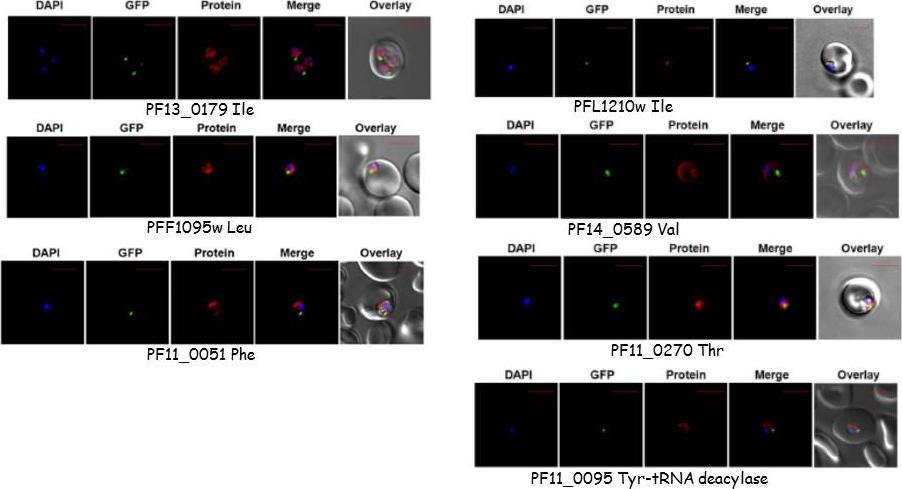
Targeting of P. falciparum aminoacyl-tRNA synthetase and of DTD. The parasite line used was GFP-tagged (strain D10 ACPleader-GFP) where apicoplast fluorescence is in green. DAPI staining is in blue while aminoacyl-tRNA synthetases are stained with Alexa594 (red). only the late ring and early trophozoite-stage localizations are depicted here but patterns were identical in other intra-erythrocytic stages. All tRNA synthases were cytoplasmic except PFL1210w Ile which is seen located in the apicoplast and deacylase which is mitochondrial.Khan S, Sharma A, Jamwal A, Sharma V, Pole AK, Thakur KK, Sharma A. Uneven spread of cis- and trans-editing aminoacyl-tRNA synthetase domains within translational compartments of P. falciparum. Sci Rep. 2011;1:188.
See original on MMP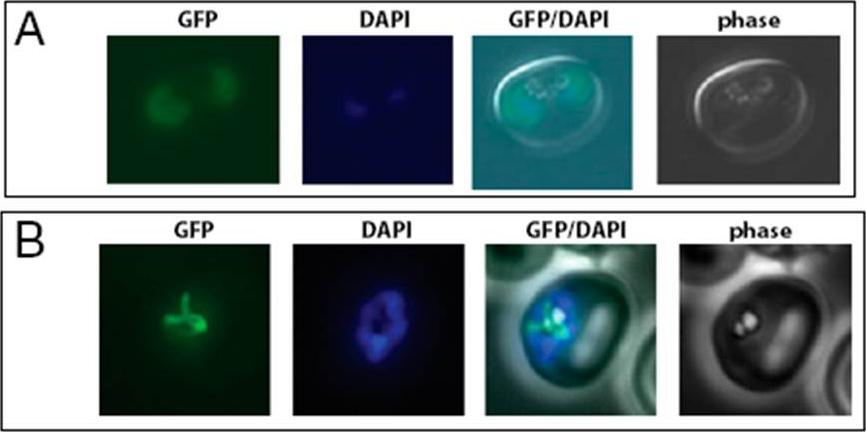
Localization of two tRNAIle: PF13_0179 is cytoplasmic, PFL1210w is in the apicoplast. (A) Live fluorescence microscopy of a doubly infected RBC expressing C-terminally tagged PF13_0179. The enzyme is expressed from the native promoter. (B) Live fluorescence microscopy of a schizont-stage parasite episomally expressing the N-terminal 92 amino acid of PFL1210w fused to GFP.Istvan ES, Dharia NV, Bopp SE, Gluzman I, Winzeler EA, Goldberg DE. Validation of isoleucine utilization targets in Plasmodium falciparum. Proc Natl Acad Sci U S A. 2011 108:1627-32.
See original on MMPMore information
| PlasmoDB | PBANKA_1347700 |
| GeneDB | PBANKA_1347700 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PB001141.02.0, PB300750.00.0, PBANKA_134770 |
| Orthologs | PCHAS_1352300 , PF3D7_1332900 , PKNH_1267700 , PVP01_1221800 , PVX_082575 , PY17X_1352700 |
| Google Scholar | Search for all mentions of this gene |