PBANKA_1126800 HECT-domain (ubiquitin-transferase), putative
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. berghei ANKA | Asexual |
Attenuated |
PlasmoGEM (Barseq) | PlasmoGEM |
Imaging data (from Malaria Metabolic Pathways)
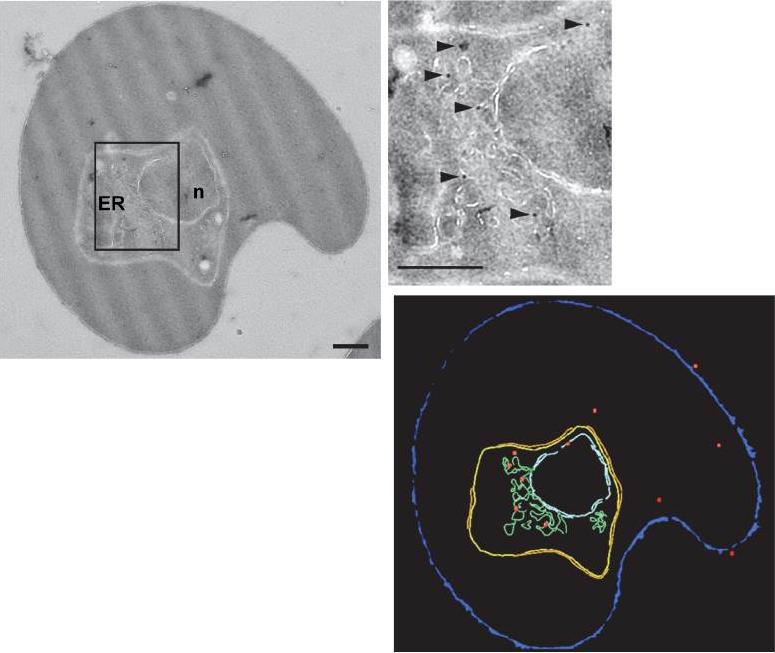
Subcellular localization of PfUT by immunoelectron microscopy. The upper panel shows a repre-sentative micrograph of a P. falciparum-infected erythrocyte preserved by high-pressure freezing and freeze-substitution, and immunolabelled with a rabbit antiserum specific to the N-terminal domain of PfUT (1:100) coupled to 10 nm protein A colloidal gold. The lower panel shows the surface rendered view of the micrograph, with red dots representing gold grains. Insert: Magnification of boxed section in micrograph. Arrowheads point towards gold label. n, nucleus; fv, food vacuole. Scale bar in D and E, 500 nm. Gold grains were significantly more present in areas of ER/Golgi complex (ER) than in other subcellular compartments.Sanchez CP, Liu CH, Mayer S, Nurhasanah A, Cyrklaff M, Mu J, Ferdig MT, Stein WD, Lanzer M. A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in Plasmodium falciparum. PLoS Genet. 2014 10(5):e1004382.
See original on MMP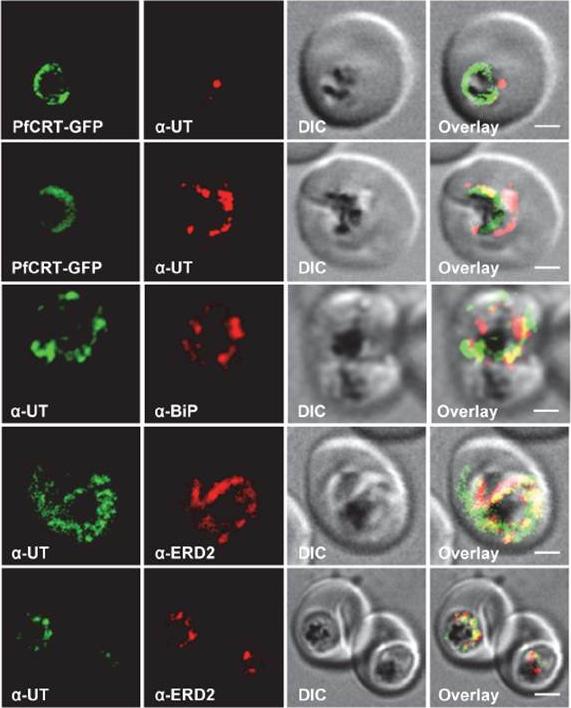
Subcellular localization of PfUT (ubiquitin-transferase). P. falciparum-infected erythrocytes at the trophozoite stage were fixed and analyzed by immunofluorescence assays using antisera to the ER marker BiP (rabbit, 1:1000), the Golgi marker ERD2 (rat, 1:500), and the N- (panels 1 and 5, rabbit, 1:3000; panel 3, mouse, 1:2000) and C-terminal domains of PfUT (panels 2 and 4, rabbit, 1:3000). Panel 1 shows a late ring stage parasite, the other panels show trophozoites. GFP fluorescence was detected, by confocal fluorescence microscopy, in parasites expressing episomally a PfCRT/GFP fusion protein. The different antisera raised against PfUT showed comparable results. Bar, 2 mm. Immunofluorescence microscopy partially co-localized PfUT with the ER marker BiP and the Golgi marker ERD2, but not with PfCRT.Sanchez CP, Liu CH, Mayer S, Nurhasanah A, Cyrklaff M, Mu J, Ferdig MT, Stein WD, Lanzer M. A HECT Ubiquitin-Protein Ligase as a Novel Candidate Gene for Altered Quinine and Quinidine Responses in Plasmodium falciparum. PLoS Genet. 2014 10(5):e1004382.
See original on MMP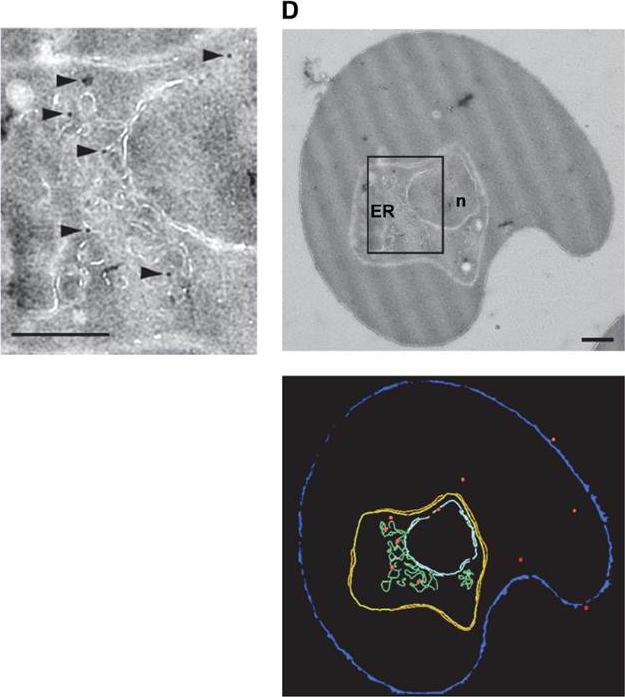
HECT ubiquitin-protein ligase (PfUT) localizes to the ER/Golgi complex. Right panels: Subcellular localization of PfUT by immunoelectron microscopy. The upper panel shows a representative micrograph of a P. falciparum-infected erythrocyte preserved by high-pressure freezing and freeze-substitution, and immunolabelled with a rabbit antiserum specific to the N-terminal domain of PfUT coupled to 10 nm protein A colloidal gold. The lower panel shows the surface rendered view of the micrograph, with red dots representing gold grains. Left: Magnification of boxed section in micrograph. Arrowheads point towards gold label. n, nucleus; fv, food vacuole. Scale bar in D and E, 500 nm. Quantitative immunoelectron microscopy confirmed a predominant localization of PfUT at the ER/Golgi complex.Sanchez CP, Liu CH, Mayer S, Nurhasanah A, Cyrklaff M, Mu J, Ferdig MT, Stein WD, Lanzer M. A HECT ubiquitin-protein ligase as a novel candidate gene for altered quinine and quinidine responses in Plasmodium falciparum. PLoS Genet. 2014 10(5):e1004382.
See original on MMP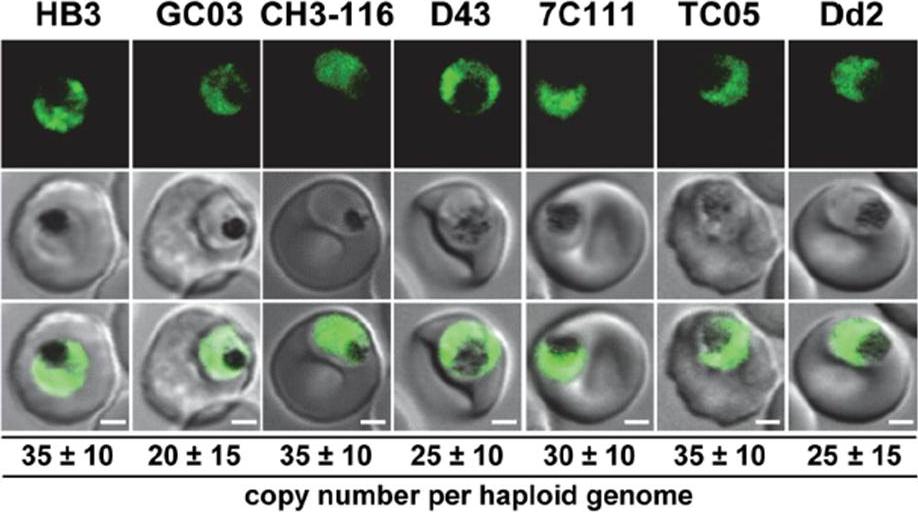
Live cell images of the P. falciparum strains indicated expressing an episomal copy of the catalytic domain of PfUT fused to GFP. The fluorescence signal is located in the parasite’s cytoplasm. Bar, 2 mM. The copy number of the plasmid per haploid genome is indicated. The means 6 SEM of at least 6 biological replicates are shown. The copy numbers are not statistically different in the transfectants. Cytoplasmic expression of the fusion protein was confirmed by fluorescence microscopy. The transfected strains maintained the vector at comparable copy numbers per haploid genome of approximately 30, with no significant differences between the transfectants.Sanchez CP, Liu CH, Mayer S, Nurhasanah A, Cyrklaff M, Mu J, Ferdig MT, Stein WD, Lanzer M. A HECT ubiquitin-protein ligase as a novel candidate gene for altered quinine and quinidine responses in Plasmodium falciparum. PLoS Genet. 2014 10(5):e1004382.
See original on MMP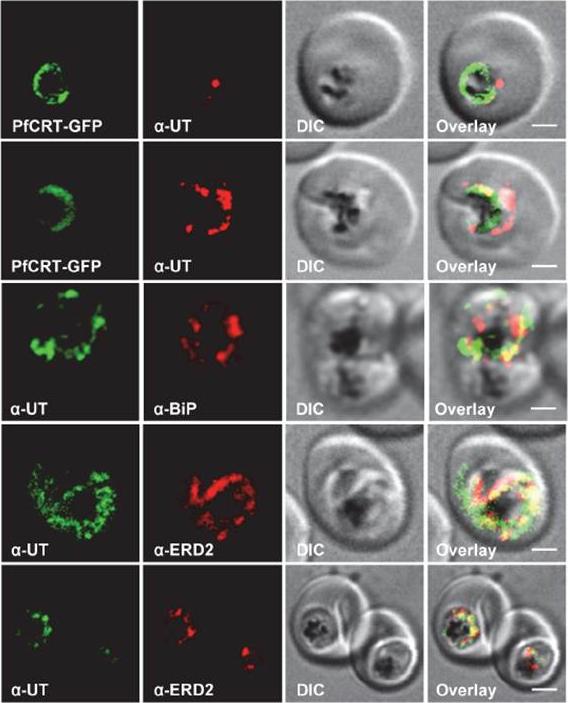
HECT ubiquitin-protein ligase (PfUT) localizes to the ER/Golgi complex. Subcellular localization of PfUT. P. falciparum-infected erythrocytes at the trophozoite stage were fixed and analyzed by immunofluorescence assays using antisera to the ER marker BiP, the Golgi marker ERD2, and the N- (panels 1 and 5, rabbit and C-terminal domains of PfUT. Panel 1 shows a late ring stage parasite, the other panels show trophozoites. GFP fluorescence was detected, by confocal fluorescence microscopy, in parasites expressing episomally a PfCRT/GFP fusion protein. The different antisera raised against PfUT showed comparable results. Bar, 2 mm. Immunofluorescence microscopy partially co-localized PfUT with the ER marker BiP and the Golgi marker ERD2, but not with PfCRT. Quantitative immunoelectron microscopy confirmed a predominant localization of PfUT at the ER/Golgi complex.Sanchez CP, Liu CH, Mayer S, Nurhasanah A, Cyrklaff M, Mu J, Ferdig MT, Stein WD, Lanzer M. A HECT ubiquitin-protein ligase as a novel candidate gene for altered quinine and quinidine responses in Plasmodium falciparum. PLoS Genet. 2014 10(5):e1004382.
See original on MMPMore information
| PlasmoDB | PBANKA_1126800 |
| GeneDB | PBANKA_1126800 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PB000952.03.0, PB100015.00.0, PB100016.00.0, PB103499.00.0, PB103867.00.0, PB105259.00.0, PB105704.00.0, PB105705.00.0, PB107195.00.0, PB108086.00.0, PB402772.00.0, PB402773.00.0, PB404039.00.0, PB404897.00.0, PBANKA_112680 |
| Orthologs | PCHAS_1126300 , PF3D7_0628100 , PKNH_1121700 , PVP01_1121400 , PVX_114510 , PY17X_1128300 |
| Google Scholar | Search for all mentions of this gene |