PBANKA_1017500 lipase, putative (UIS28)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Possible |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Possible |
18614010 | Theo Sanderson, Wellcome Trust Sanger Institute | |
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
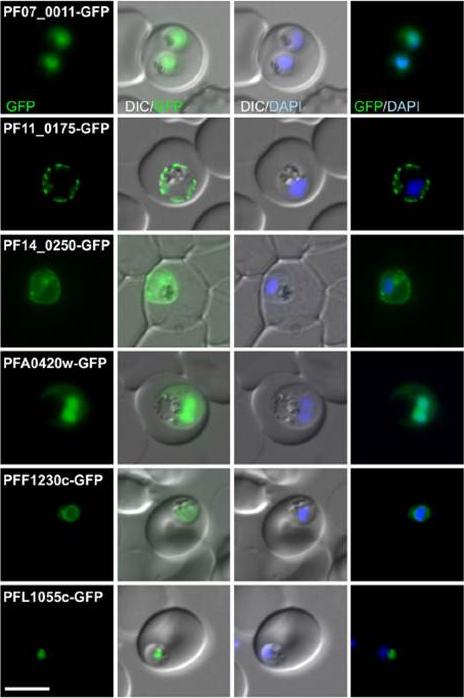
Fluorescence pattern of non-exported (B) or exported (C) GFP fusion proteins. PF07_0011 . unknown function; PF11_0175 heat shock protein 101; PF14_0250 lipase; PFA0429w unknown function; PFF1230c unknown function; PFL1055c unknown function. PF07_0011 and PFA0420w were in the nucleus with a cytoplasmic pool, PF11_0175 and PF14_0250 were at the parasite periphery (consistent with a PPM, PV or PVM location, PFF1230c was perinuclear, suggesting an ER location, and PFL1055c was found in a nucleus proximal focus that co-localized with the GolgiHeiber A, Kruse F, Pick C, Grüring C, Flemming S, Oberli A, Schoeler H, Retzlaff S, Mesén-Ramírez P, Hiss JA, Kadekoppala M, Hecht L, Holder AA, Gilberger TW, Spielmann T. Identification of New PNEPs Indicates a Substantial Non-PEXEL Exportome and Underpins Common Features in Plasmodium falciparum Protein Export. PLoS Pathog. 2013 9(8):e1003546.
See original on MMPMore information
| PlasmoDB | PBANKA_1017500 |
| GeneDB | PBANKA_1017500 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PB000864.01.0, PB300412.00.0, PBANKA_101750, UIS28 |
| Orthologs | PCHAS_1018300 , PF3D7_1427100 , PKNH_1331600 , PVP01_1322200 , PVX_085180 , PY17X_1019000 |
| Google Scholar | Search for all mentions of this gene |