PBANKA_0941900 histone H4, putative
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Refractory |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
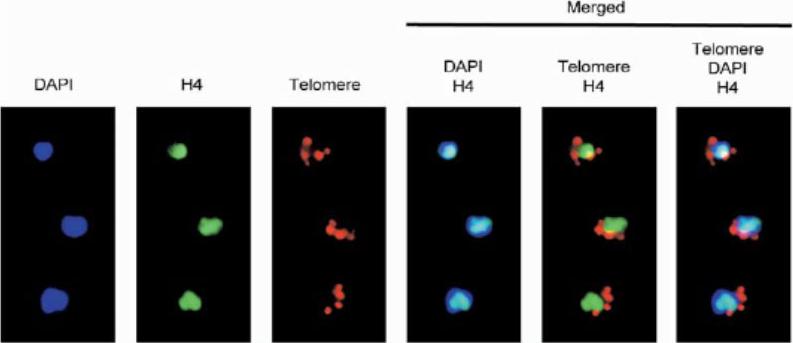
Combination of immunolocalization (antiacetylated H4 antibodies, green) and FISH (telomere, red) colocalization analysis of trophozoite-stage parasites. The nucleus is stained with DAPI. Acetylated H4 histone is excluded from the nuclear periphery.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mâncio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005 121:25-36. Copyright Elsevier 2009.
See original on MMP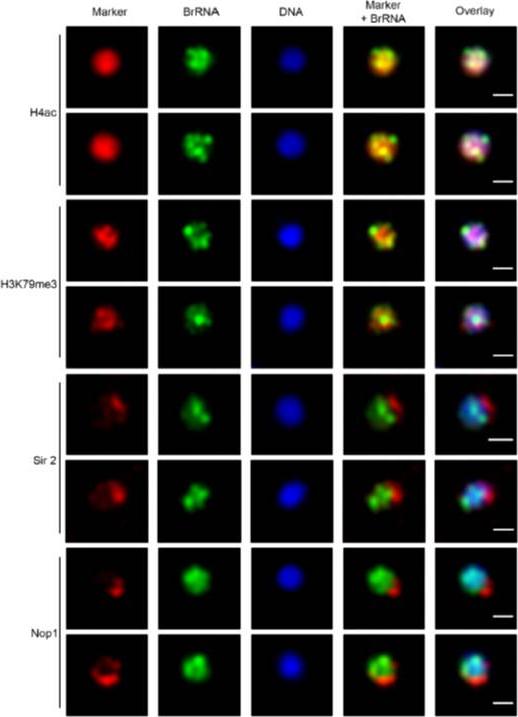
Transcription sites form a distinct compartment in the nucleus of P. falciparum. Immunofluorescence assay for nascent RNAs of ring stage parasites labeled with BrUTP (BrRNA, green) and the compartments of acetylated H4 (H4ac, active chromatin); a proposed histone marker of nascent transcription (H3K79me3; PfSir2A (silencing compartment); and Nop1 (nucleolar compartment). DNA was labeled by DAPI, in blue. Bars, 1 mm.Moraes CB, Dorval T, Contreras-Dominguez M, Dossin Fde M, Hansen MA, Genovesio A, Freitas-Junior LH. Transcription sites are developmentally regulated during the asexual cycle of Plasmodium falciparum. PLoS One. 2013;8(2):e55539.
See original on MMP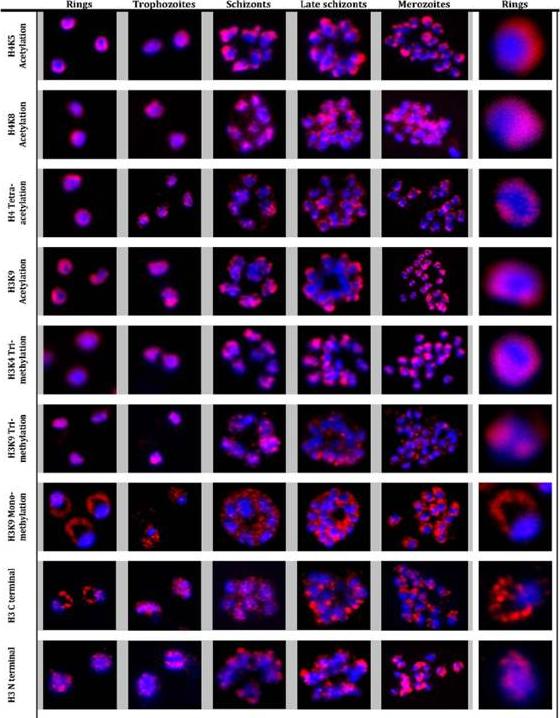
Immunofluorescence analysis of histone modifications in P. falciparum. Localization of histone modifications were analyzed with the ring, trophozoite, schizont, late schizont and merozoite stages of the IDC. IFAs were carried out with antibodies against specific histone 3 and 4 lysine residue acetylations: H3K9Ac, H4K5Ac, H4K8Ac, and H4Ac4, as well as methylations: H3K4Me3, H3K9Me3 and H3K9Me1, and unmodified histone H3. Nuclear DNA was stained with DAPI (blue). All modifications, with the exception of H3K9Me1 and H3 (antibody raised against H3 C terminal), showed specific and distinct localization in the nucleus in all the stages. In contrast, H3K9Me1 was localized mainly outside the nucleus with very low levels detected inside the nucleus. Luah YH, Chaal BK, Ong EZ, Bozdech Z. A moonlighting function of Plasmodium falciparum histone 3, mono-methylated at lysine 9? PLoS One. 2010 5(4):e10252.
See original on MMP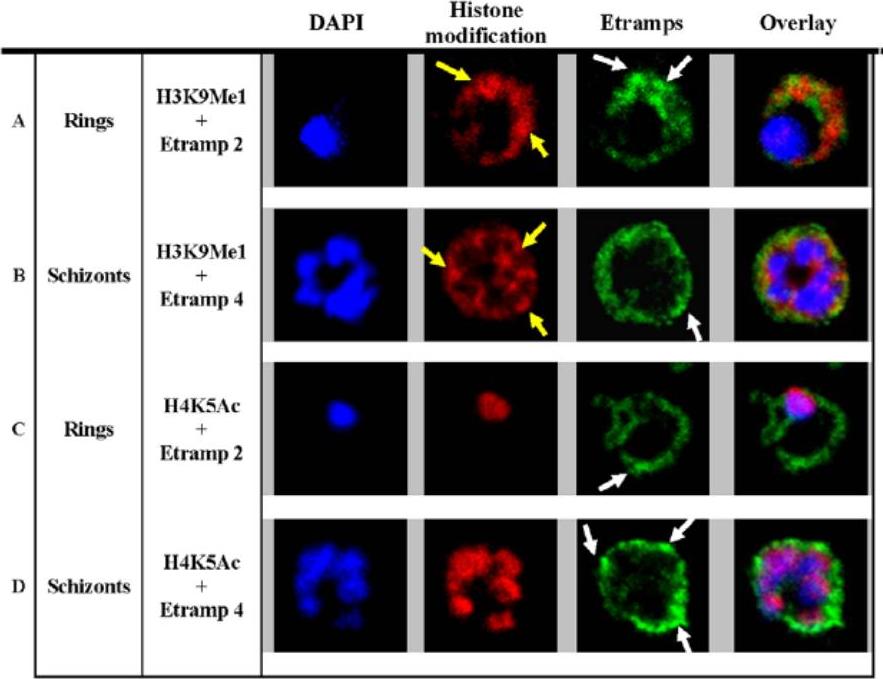
H3K9Me1 localized to the parasitophorous vacuole during the ring stage. Co-localization of H3K9Me1 with Etramp 2 (A) and 4 (B) was performed in ring and schizont stage parasites respectively. Similarly, co-localization of H4K5Ac with Etramp 2 (C) and 4 (D) was performed in ring and schizont stage parasites respectively. H3K9Me1/H4K5Ac and Etramp 2/4 were stained red and green respectively. DAPI stained nuclear DNA blue. Yellow and white arrows indicate foci of more intense fluorescence produced by H3K9Me1 and Etramp labeling respectively. In ring stage parasites, compared to schizonts, H3K9Me1 partially co-localized with Etramp 2 indicating localization to different compartments of the PV. H4K5Ac was localized solely to the nucleus and did not co-localize with either Etramp 2 or 4.Luah YH, Chaal BK, Ong EZ, Bozdech Z. A moonlighting function of Plasmodium falciparum histone 3, mono-methylated at lysine 9? PLoS One. 2010 5(4):e10252.
See original on MMP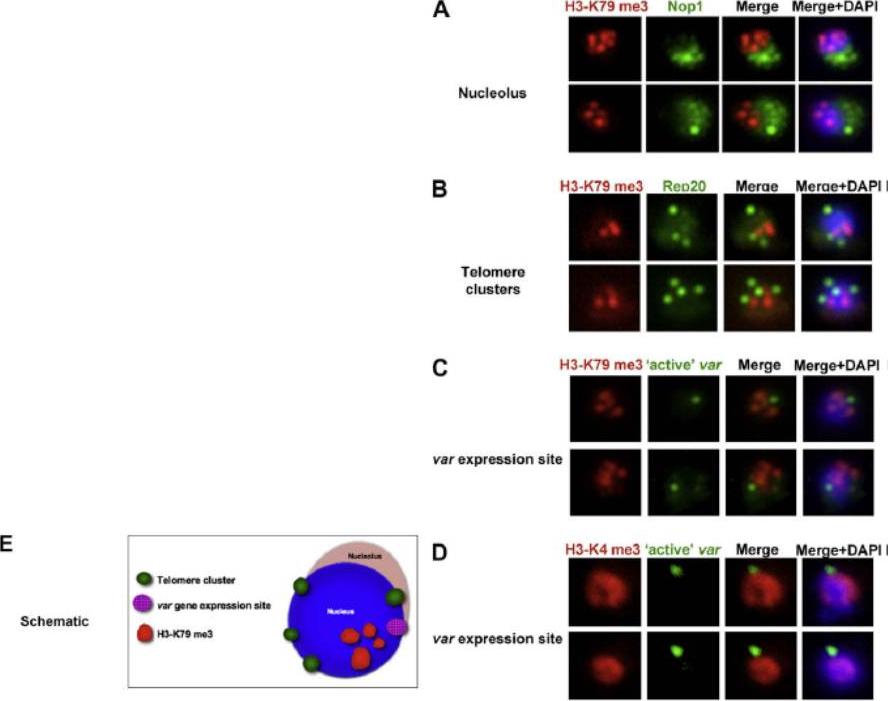
Co-localisation of H3-K79me3 with known cellular compartments. A. Nucleolus: Double labelling IFAs with antibodies against H3-K79me3 and nucleolus marker, PfNop1. B. Telomere clusters: IFeFISH analysis between H3-K79me3 (red) and Rep20 (green). C. Active var gene: IF-FISH analysis between H3-K79me3 (red) and active var gene (green). D. Active var gene colocalisation with activating mark, H3-K4me3. IF-FISH analysis between H3-K4me3 (red) and active var gene (green). DAPI stained nucleus in blue. Font colour represents corresponding fluorescein labelled antibody. Scale bar depicts 1 mm. E. Schematic of the dynamic compartmen-talisation of the P. falciparum nucleus. The distinct sub-nuclear distribution of histone methylation modification, taking example of H3-K79me3 as observed in this study. Known compartments compatible with active transcription are indicated. H3-K79me3 occupies a distinct zone.Issar N, Ralph SA, Mancio-Silva L, Keeling C, Scherf A. Differential sub-nuclear localisation of repressive and activating histone methyl modifications in P. falciparum. Microbes Infect. 2009 11:403-7.
See original on MMPMore information
| PlasmoDB | PBANKA_0941900 |
| GeneDB | PBANKA_0941900 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PBANKA_094190 |
| Orthologs | PCHAS_0902400 , PF3D7_1105000 , PKNH_0902600 , PVP01_0905800 , PVX_090930 , PY17X_0944400 |
| Google Scholar | Search for all mentions of this gene |