PBANKA_0923800 pyruvate dehydrogenase E1 component subunit alpha (pdhA)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Possible |
RMgm-854 | Imported from RMgmDB | |
| P. berghei ANKA |
Possible |
RMgm-677 | Imported from RMgmDB | |
| P. berghei ANKA |
Refractory |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. yoelii yoelii 17X |
Possible |
RMgm-378 | Imported from RMgmDB | |
| P. yoelii yoelii 17X |
Possible |
RMgm-376 | Imported from RMgmDB | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. berghei ANKA | Asexual |
No difference |
RMgm-854 | Imported from RMgmDB |
| P. berghei ANKA | Asexual |
No difference |
RMgm-677 | Imported from RMgmDB |
| P. berghei ANKA | Gametocyte |
No difference |
RMgm-677 | Imported from RMgmDB |
| P. berghei ANKA | Ookinete |
No difference |
RMgm-677 | Imported from RMgmDB |
| P. berghei ANKA | Oocyst |
No difference |
RMgm-854 | Imported from RMgmDB |
| P. berghei ANKA | Oocyst |
No difference |
RMgm-677 | Imported from RMgmDB |
| P. berghei ANKA | Sporozoite |
No difference |
RMgm-854 | Imported from RMgmDB |
| P. berghei ANKA | Sporozoite |
No difference |
RMgm-677 | Imported from RMgmDB |
| P. berghei ANKA | Liver |
Difference from wild-type |
RMgm-854
Strongly reduced infectivity of sporozoites. Only a very low number of mice developed blood stage parasitemia after infection with high numbers of sporozoites.Maturing liver stages abort development as a result of a combination of the lack of expression of PDH-E1 and expression of FPO (see additional information) |
Imported from RMgmDB |
| P. berghei ANKA | Liver |
Difference from wild-type |
RMgm-677
See 'Additional remarks phenotype'. Partly aberrant formation of mature liver-stage schizonts |
Imported from RMgmDB |
| P. yoelii yoelii 17X | Asexual |
No difference |
RMgm-378 | Imported from RMgmDB |
| P. yoelii yoelii 17X | Asexual |
No difference |
RMgm-376 | Imported from RMgmDB |
| P. yoelii yoelii 17X | Gametocyte |
No difference |
RMgm-378 | Imported from RMgmDB |
| P. yoelii yoelii 17X | Gametocyte |
No difference |
RMgm-376 | Imported from RMgmDB |
| P. yoelii yoelii 17X | Ookinete |
No difference |
RMgm-378 | Imported from RMgmDB |
| P. yoelii yoelii 17X | Ookinete |
No difference |
RMgm-376 | Imported from RMgmDB |
| P. yoelii yoelii 17X | Oocyst |
No difference |
RMgm-378 | Imported from RMgmDB |
| P. yoelii yoelii 17X | Oocyst |
No difference |
RMgm-376 | Imported from RMgmDB |
| P. yoelii yoelii 17X | Sporozoite |
No difference |
RMgm-378 | Imported from RMgmDB |
| P. yoelii yoelii 17X | Sporozoite |
No difference |
RMgm-376 | Imported from RMgmDB |
| P. yoelii yoelii 17X | Liver |
Difference from wild-type |
RMgm-378
BALB/c mice intravenously injected with 1x10(4)-5x10(4) salivary gland sporozoites did not develop blood stage infections. Analysis of in vitro cultured liver stages showed significant defects in terms of growth and nuclear division. At 24 h pi, no difference was observed between mutant and wild type liver stages with regard to protein expression and parasite size. During late liver stage development (43 and 52 h pi), no expression was detected of the merozoite-specific protein MSPI in mutant parasites. Mutant parasites showed significant defects in terms of growth and nuclear division and the average size of the mutant parasites at 43 h pi was approximately 30% of wild type parasites. |
Imported from RMgmDB |
| P. yoelii yoelii 17X | Liver |
Difference from wild-type |
RMgm-376
BALB/c mice intravenously injected with 1x10(4)-5x10(4) salivary gland sporozoites did not develop blood stage infections. Analysis of in vitro cultured liver stages showed significant defects in terms of growth and nuclear division. At 24 h pi, no difference was observed between mutant and wild type liver stages with regard to protein expression and parasite size. During late liver stage development (43 and 52 h pi), no expression was detected of the merozoite-specific protein MSPI in mutant parasites. Mutant parasites showed significant defects in terms of growth and nuclear division and the average size of the mutant parasites at 43 h pi was approximately 30% of wild type parasites. |
Imported from RMgmDB |
Imaging data (from Malaria Metabolic Pathways)
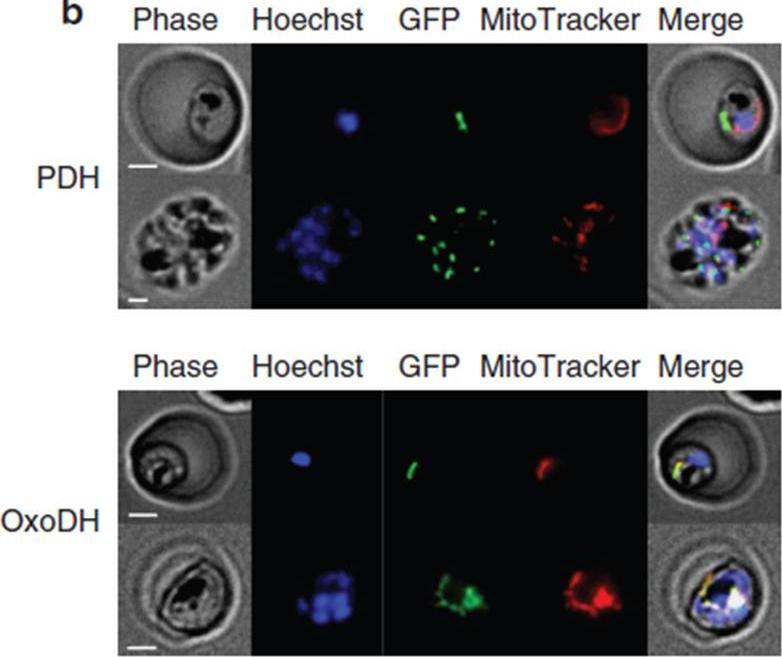
Localization of PfPDH and PfOxoDH within erythrocytes infected with 3D7 parasites. Localization of PfPDH (top panel) and PfOxoDH (bottom panel) in transgenic parasites. The left column shows phase contrast images, followed by fluorescence images of a nuclear dye (Hoechst; blue), GFP-tagged leader sequences of the target enzymes (green), a mitochondrial dye MitoTracker; red) and all four images merged together (right column). Scale bars, 2 mm. The E1-a subunit of PfPDH localizes to a discrete organelle adjacent to the mitochondrion and distinct from the nucleus, consistent with an apicoplast localization (top panel). The truncated E1 subunit of PfOxoDH, which has been predicted to have a mitochondrial localization,colocalizes with MitoTracker when fused to GFP (bottom panel), consistent with a mitochondrial localization.Chan XW, Wrenger C, Stahl K, Bergmann B, Winterberg M, Müller IB, Saliba KJ. Chemical and genetic validation of thiamine utilization as an antimalarial drug target. Nat Commun. 2013 Jun 27;4:2060.
See original on MMP
N-terminal targeting sequences of PDH subunits E1a and E2 of P. falciparum direct GFP to the apicoplast in intraerythrocytic parasites. The panels show images of P. falciparum parasites from different blood stages expressing episomally encoded GFP fusion proteins. Colocalization with the apicoplast-resident protein ACP using a-ACP antibodies unambiguously shows that the pre-sequences of PDH subunits E1a and E2 are sufficient to target proteins exclusively to the apicoplast (A and C). Red MitoTracker labelling confirms that the GFP staining is for the apicoplast – distinct from but in close proximity to the mitochondrion (B and D). Note also the characteristic shape of the apicoplast in the different parasite.Foth BJ, Stimmler LM, Handman E, Crabb BS, Hodder AN, McFadden GI. The malaria parasite Plasmodium falciparum has only one pyruvate dehydrogenase complex, which is located in the apicoplast. Mol Microbiol. 2005 55(1):39-53.
See original on MMPMore information
| PlasmoDB | PBANKA_0923800 |
| GeneDB | PBANKA_0923800 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PB301288.00.0, PBANKA_092380 |
| Orthologs | PCHAS_0920600 , PF3D7_1124500 , PKNH_0922300 , PVP01_0925100 , PVX_091840 , PY17X_0925800 |
| Google Scholar | Search for all mentions of this gene |