PBANKA_0912300 tudor staphylococcal nuclease, putative (TSN)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Possible |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. berghei ANKA | Asexual |
No difference |
PlasmoGEM (Barseq) | PlasmoGEM |
Imaging data (from Malaria Metabolic Pathways)
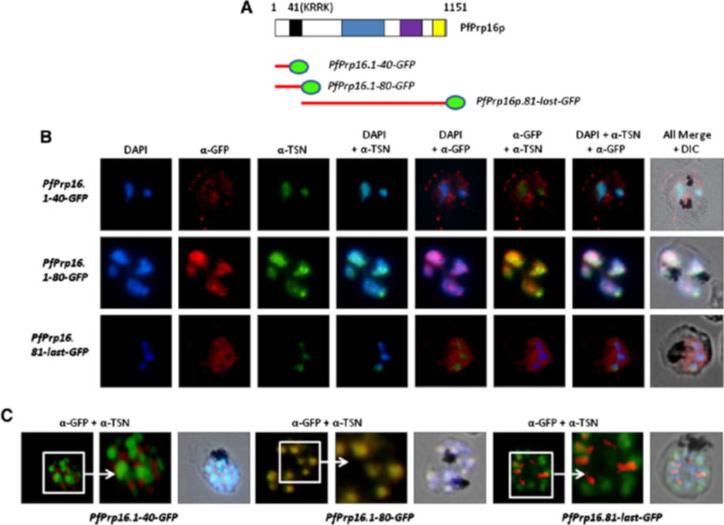
N-terminal domain of the PfPrp16p targets the protein to the nucleus. (A) Schematic representation of the PfPrp16-GFP chimeric constructs. Predicted nuclear localization sequence, “KRRK,” is shown as a black box at 41st–44th amino acid position. (B and C) Immunofluorescence assay based localization of the chimeric GFP proteins at (B) trophozoite and (C) schizont stages respectively. Anti-GFP (red, 1:200 dilution) and anti-PfTSN (green, 1:100 dilution) antibodies were used on thin blood smears of parasite lines. PfPrp16.1-40-GFP and PfPrp16.81-last-GFP chimeric proteins do not localize with the nuclear marker PfTSN protein in either of the developmental stages, whereas chimeric protein PfPrp16.1-80-GFP was trafficked to the nucleus in trophozoite and schizont stages.Singh PK, Kanodia S, Dandin CJ, Vijayraghavan U, Malhotra P. Plasmodium falciparum Prp16 homologue and its role in splicing. Biochim Biophys Acta. 2012 1819(11-12):1186-99
See original on MMP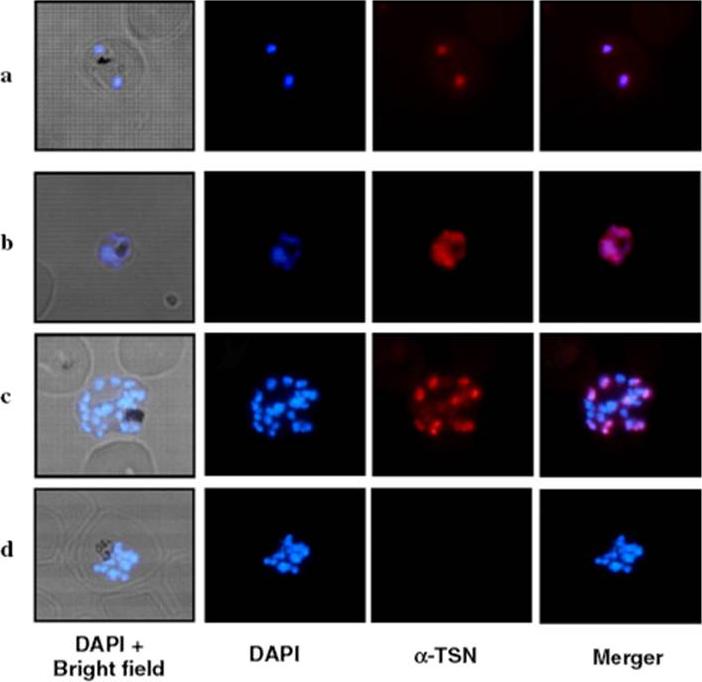
Expression of PfTSN in asexual blood stages of P. falciparum. (A) Immunofluorescence assay performed using anti-PfDTSN antisera (1:200 dilution) on thin blood smears of P. falciparum. PfTSN protein localized mainly in the nucleus in all three asexual blood stages, ring (a), trophozoite (b) and schizont (c) stages. DAPI staining was used to localize the P. falciparum nucleus. A pre-immune serum (1:50 dilution) was used as a control (d).Hossain MJ, Korde R, Singh S, Mohmmed A, Dasaradhi PV, Chauhan VS, Malhotra P. Tudor domain proteins in protozoan parasites and characterization of Plasmodium falciparum tudor staphylococcal nuclease. Int J Parasitol. 2008 38:513-26 Copyright Elsevier
See original on MMP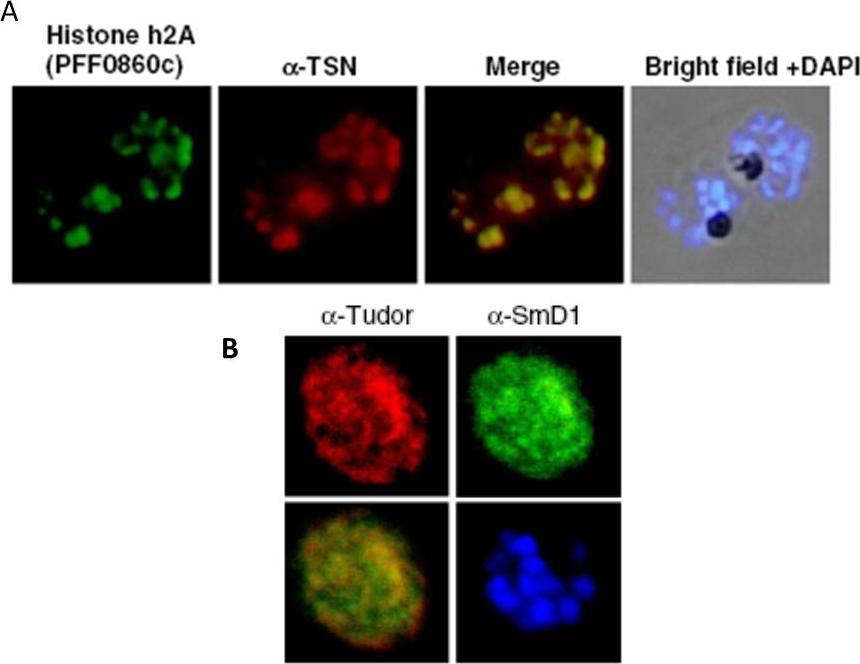
Upper panel: Co-localization of PfTudor-SN and Histone h2A. Immunofluorescence assays were performed at P. falciparum schizont stage using peptide antibodies generated against Histone h2A. The interacting h2A in the parasite were co-localized with anti-PfTudor-SN antibody in the nucleus.Lower panel: .Co-localization of PfTudor-SN and SmD1 protein of the parasite. PfTudor-SN was also co-localized with the PfSmD1 suggesting a close association between themHossain MJ, Korde R, Singh PK, Kanodia S, Ranjan R, Ram G, Kalsey GS, Singh R, Malhotra P. Plasmodium falciparum Tudor Staphylococcal Nuclease interacting proteins suggest its role in nuclear as well as splicing processes. Gene. 2010 468:48-57. Copyright Elsevier 2011
See original on MMP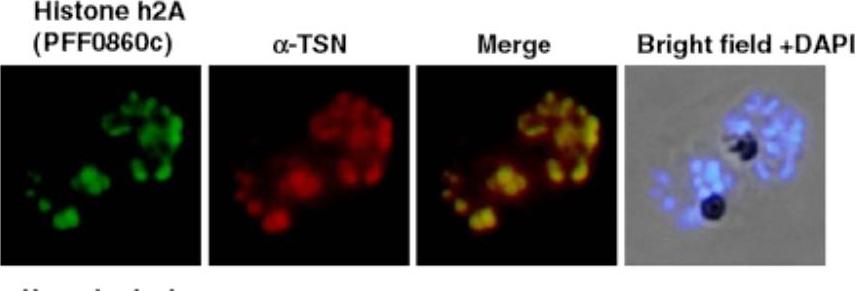
Co-localization of PfTudor-SN and histone H2A. The interacting proteins in the parasite were co-localized with anti-PfTudor-SN antibody. FITC and Cy3 labeled secondary antibodies were used for H2A and anti-TSN, respectively. Both proteins are mainly localized in the nucleus.Hossain MJ, Korde R, Singh PK, Kanodia S, Ranjan R, Ram G, Kalsey GS, Singh R, Malhotra P. Plasmodium falciparum Tudor Staphylococcal Nuclease interacting proteins suggest its role in nuclear as well as splicing processes. Gene. 2010 468(1-2):48-57. PMID:
See original on MMP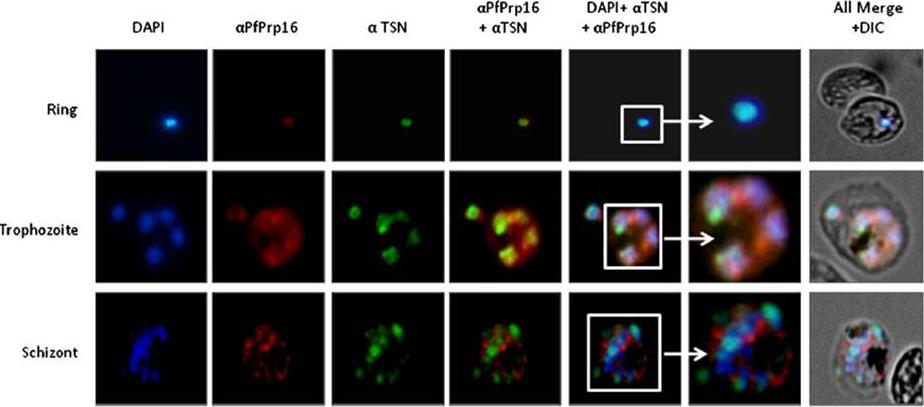
Expression of the PfPrp16p during asexual blood stages of P. falciparum. Immunofluorescence assay based localization of the PfPrp16p. Anti-PfPrp16p sera (1:200 dilution) were used to localized the protein using thin blood smears of P. falciparum ring, trophozoite and schizont stages. DAPI and anti-Pf‐Tudor-SN (PfTSN) antibody were used to localize the nucleus. PfTSN co-localizes with Pf-Tudor-SN (PfTSN), a nuclear resident protein. At the trophozoite and schizont stages the protein was detected in the nucleus as well as in the cytoplasm indicating nucleo-cytoplasmic shuttling of PfPrp16p.Singh PK, Kanodia S, Dandin CJ, Vijayraghavan U, Malhotra P. Plasmodium falciparum Prp16 homologue and its role in splicing. Biochim Biophys Acta. 2012 1819(11-12):1186-99
See original on MMPMore information
| PlasmoDB | PBANKA_0912300 |
| GeneDB | PBANKA_0912300 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PB001586.02.0, PBANKA_091230 |
| Orthologs | PCHAS_0713600 , PF3D7_1136300 , PKNH_0934500 , PVP01_0937200 , PVX_092430 , PY17X_0913800 |
| Google Scholar | Search for all mentions of this gene |