PBANKA_0816000 type II NADH:ubiquinone oxidoreductase (NDH2)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Possible |
RMgm-630 | Imported from RMgmDB | |
| P. berghei ANKA |
Possible |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Possible |
30964863 Normal growth rate. 'Deletion of PfNDH2 does not alter the parasite's susceptibility to multiple mtETC inhibitors, including atovaquone and ELQ-300.' |
Theo Sanderson, Google AI | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. berghei ANKA | Asexual |
No difference |
RMgm-630 | Imported from RMgmDB |
| P. berghei ANKA | Asexual |
No difference |
PlasmoGEM (Barseq) | PlasmoGEM |
| P. berghei ANKA | Gametocyte |
No difference |
RMgm-630 | Imported from RMgmDB |
| P. berghei ANKA | Ookinete |
No difference |
RMgm-630 | Imported from RMgmDB |
| P. berghei ANKA | Oocyst |
Difference from wild-type |
RMgm-630
Normal numbers of ookinetes are produced. Ookinetes showed normal (gliding) motility. Normal numbers of oocysts are produced. Mutant oocysts were considerably smaller than wild type oocysts. No sporozoite formation inside oocysts |
Imported from RMgmDB |
| P. berghei ANKA | Sporozoite |
Difference from wild-type |
RMgm-630
Normal numbers of oocysts are produced. Mutant oocysts were considerably smaller than wild type oocysts. No sporozoite formation inside oocysts. No salivary gland sporozoites. |
Imported from RMgmDB |
| P. falciparum 3D7 | Asexual |
No difference |
30964863 Normal growth rate. 'Deletion of PfNDH2 does not alter the parasite's susceptibility to multiple mtETC inhibitors, including atovaquone and ELQ-300.' |
Theo Sanderson, Google AI |
Imaging data (from Malaria Metabolic Pathways)
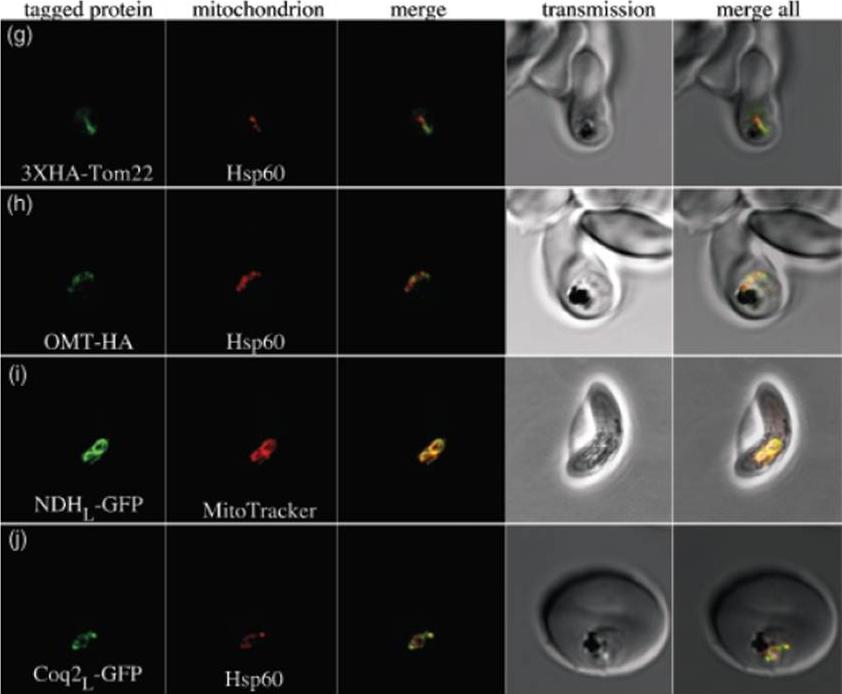
g) PfTom22 localizes to the mitochondrion of P. falciparum. Pf Tom22 was tagged with a 3X-HA epitope tag at its N-terminus. An immuno-fluorescence assay was performed with 3XHA-PfTom22 labelled in green and the mitochondrial marker Hsp60 labelled in red. The merged image (central panel) shows apparent overlap between the two proteins, indicating that PfTom22 is a mitochondrial protein. h) The oxoglutarate/malate transporter homologue of P. falciparum (PfOMT) localizes to the mitochondrion of P. falciparum. PfOMTwas tagged with a 1XHA tag at its C-terminus (green) and co-labelled with anti-Hsp60 antibody (red). The merged image shows apparent overlap between the two proteins, indicating that PfOMT is a mitochondrial protein. i) The single-subunit NAD(P)H dehydrogenase homologue of P. falciparum (PfNDH) localizes to the mitochondrion. The N-terminal extension of PfNDH was fused to green fluorescent protein (GFP, green), with live parasites co-labelled with MitoTracker Red (red), a mitochondrial marker. In this gametocyte image, there is clear overlap, indicating that PfNDH localizes to the mitochondrion. In asexual stages, when protein expression levels are probably higher, a considerable portion of PfNDHL-GFP also localizes to the cytosol, suggesting that import of this fusion protein is somewhat inefficient. j) The coenzyme Q biosynthesis protein Coq2 homologue of P. falciparum (PfCoq2) localizes to the mitochondrion. The N-terminal portion of PfCoq2 was fused to green fluorescent protein and labelled with anti-green fluorescent protein antibody (green). These fixed parasites were co-labelled with anti-Hsp60 antibody (red), with the apparent co-localization indicating that PfCoq2 is a mitochondrial proteinvan Dooren GG, Stimmler LM, McFadden GI. Metabolic maps and functions of the Plasmodium mitochondrion. FEMS Microbiol Rev. 2006 30(4):596-630. Review.
See original on MMPMore information
| PlasmoDB | PBANKA_0816000 |
| GeneDB | PBANKA_0816000 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PB300677.00.0, PB301156.00.0, PBANKA_081600 |
| Orthologs | PCHAS_0816300 , PF3D7_0915000 , PKNH_0713000 , PVP01_0713400 , PVX_099180 , PY17X_0819300 |
| Google Scholar | Search for all mentions of this gene |