PBANKA_0610100 lysine-specific histone demethylase 1, putative (LSD1)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Possible |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
None reported yet. Please press the '+' button above to add one.Imaging data (from Malaria Metabolic Pathways)
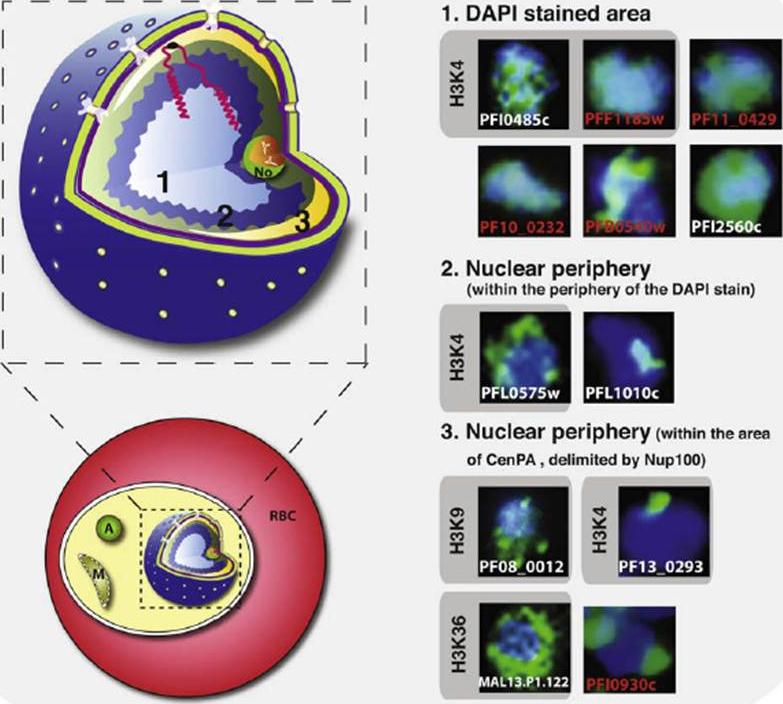
Schematic representation of the Plasmodium falciparum nuclear architecture. A schematic representation of an infected red blood cell (RBC) and an enlargement of the P. falciparum nucleus are shown. From the centre towards the periphery we have defined three distinct areas (1–3) in which nuclear proteins specifically distribute. A summary is shown in which trophozoite stages have been analyzed. Proteins, previously predicted to functionally interact, are shown in red. Proteins previously predicted for their histone target sites are indicated by grey boxes. A, apicoplast. M, mitochondria, No, nucleolus.Volz J, Carvalho TG, Ralph SA, Gilson P, Thompson J, Tonkin CJ, Langer C, Crabb BS, Cowman AF. Potential epigenetic regulatory proteins localize to distinct nuclear sub-compartments in Plasmodium falciparum. Int J Parasitol. 2010 40(1):109-21. Copyright Elsevier.
See original on MMPMore information
| PlasmoDB | PBANKA_0610100 |
| GeneDB | PBANKA_0610100 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PB000565.00.0, PB000942.02.0, PB001066.01.0, PB105917.00.0, PB300881.00.0, PBANKA_061010 |
| Orthologs | PCHAS_0611800 , PF3D7_1211600 , PKNH_1311700 , PVP01_1310800 , PVX_084610 , PY17X_0612600 |
| Google Scholar | Search for all mentions of this gene |