PBANKA_0604700 shewanella-like protein phosphatase 2, putative (SHLP2)
Disruptability [+]
| Species | Disruptability | Reference | Submitter | |
|---|---|---|---|---|
| P. berghei ANKA |
Possible |
RMgm-1036 | Imported from RMgmDB | |
| P. berghei ANKA |
Possible |
PlasmoGEM (Barseq) | PlasmoGEM | |
| P. falciparum 3D7 |
Possible |
29073264 | Theo Sanderson, Wellcome Trust Sanger Institute | |
| P. falciparum 3D7 |
Possible |
USF piggyBac screen (Insert. mut.) | USF PiggyBac Screen | |
Mutant phenotypes [+]
| Species | Stage | Phenotype | Reference | Submitter |
|---|---|---|---|---|
| P. berghei ANKA | Asexual |
No difference |
RMgm-1036 | Imported from RMgmDB |
| P. berghei ANKA | Asexual |
No difference |
PlasmoGEM (Barseq) | PlasmoGEM |
| P. berghei ANKA | Gametocyte |
No difference |
RMgm-1036 | Imported from RMgmDB |
| P. berghei ANKA | Ookinete |
No difference |
RMgm-1036 | Imported from RMgmDB |
| P. berghei ANKA | Oocyst |
No difference |
RMgm-1036 | Imported from RMgmDB |
| P. berghei ANKA | Sporozoite |
No difference |
RMgm-1036 | Imported from RMgmDB |
| P. berghei ANKA | Liver |
No difference |
RMgm-1036 | Imported from RMgmDB |
| P. falciparum 3D7 | Asexual |
No difference |
29073264 | Theo Sanderson, Wellcome Trust Sanger Institute |
Imaging data (from Malaria Metabolic Pathways)
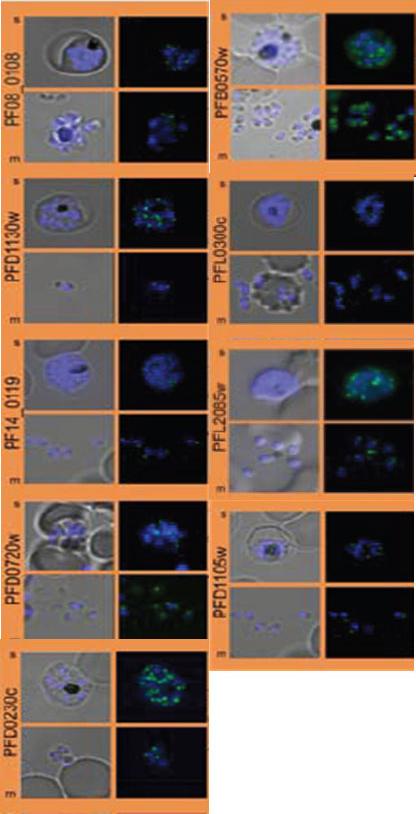
Subcellular distribution of proteins predicted to be involved in invasion. All proteins were localized in schizonts (s) and free merozoites (m) using GFP-fusion proteins and grouped according to their predominant GFP localization. Apical. Nuclei were stained with DAPI (blue).Hu G, Cabrera A, Kono M, Mok S, Chaal BK, Haase S, Engelberg K, Cheemadan S, Spielmann T, Preiser PR, Gilberger TW, Bozdech Z. Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nat Biotechnol. 2010 28(1):91-8.
See original on MMPMore information
| PlasmoDB | PBANKA_0604700 |
| GeneDB | PBANKA_0604700 |
| Malaria Metabolic Pathways | Localisation images Pathways mapped to |
| Previous ID(s) | PB001090.02.0, PB001550.02.0, PBANKA_060470 |
| Orthologs | PCHAS_0606500 , PF3D7_1206000 , PKNH_1305700 , PVP01_1305100 , PVX_084335 , PY17X_0607200 |
| Google Scholar | Search for all mentions of this gene |